Discovery of a distinct domain in cyclin A sufficient for centrosomal localization independently of Cdk binding
- PMID: 20133761
- PMCID: PMC2840291
- DOI: 10.1073/pnas.0914874107
Discovery of a distinct domain in cyclin A sufficient for centrosomal localization independently of Cdk binding
Abstract
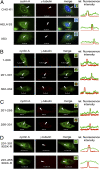
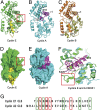
Centrosomes have recently emerged as key regulators of the cell cycle. The G1/S transition requires a functional centrosome, and centrosomal localization of numerous proteins, including cyclin/Cdk complexes, is important for the G2/M transition. Here we identify a modular centrosomal localization signal (CLS) localizing cyclin A to centrosomes independently of Cdk binding. The cyclin A CLS is located in a distinct part of the molecule compared with the cyclin E CLS and includes the MRAIL hydrophobic patch involved in substrate recognition. The cyclin A CLS interacts with p27(KIP1), and expression of p27(KIP1) removes cyclin A but not cyclin E from centrosomes. Expression of the cyclin A CLS displaces both endogenous cyclin A and E from centrosomes and inhibits DNA replication, supporting an emerging concept that DNA replication is linked to centrosomal events. Structural analysis indicates that differences in surface charge and length of the C-terminal helix explain why the MRAIL region in cyclin E is not a functional CLS. These results indicate that the cyclin A CLS may contribute to targeting and recognition of centrosomal Cdk substrates and is required for specific effects of p27(KIP1) on cyclin A-Cdk2.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
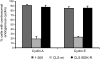
Similar articles
-
The cyclin A centrosomal localization sequence recruits MCM5 and Orc1 to regulate centrosome reduplication.J Cell Sci. 2010 Aug 15;123(Pt 16):2743-9. doi: 10.1242/jcs.073098. Epub 2010 Jul 27. J Cell Sci. 2010. PMID: 20663915 Free PMC article.
-
Centrosomal localization of cyclins E and A: structural similarities and functional differences.Cell Cycle. 2011 Jan 15;10(2):199-205. doi: 10.4161/cc.10.2.14444. Epub 2011 Jan 15. Cell Cycle. 2011. PMID: 21217199 Free PMC article.
-
Centrosomal localization of cyclin E-Cdk2 is required for initiation of DNA synthesis.Curr Biol. 2010 May 11;20(9):856-60. doi: 10.1016/j.cub.2010.03.028. Epub 2010 Apr 22. Curr Biol. 2010. PMID: 20399658 Free PMC article.
-
Molecular basis for the specificity of p27 toward cyclin-dependent kinases that regulate cell division.J Mol Biol. 2005 Jun 17;349(4):764-73. doi: 10.1016/j.jmb.2005.04.019. Epub 2005 Apr 26. J Mol Biol. 2005. PMID: 15890360
-
Regulation of cell division by intrinsically unstructured proteins: intrinsic flexibility, modularity, and signaling conduits.Biochemistry. 2008 Jul 22;47(29):7598-609. doi: 10.1021/bi8006803. Biochemistry. 2008. PMID: 18627125 Free PMC article. Review.
Cited by
-
Quantitative Systems Biology to decipher design principles of a dynamic cell cycle network: the "Maximum Allowable mammalian Trade-Off-Weight" (MAmTOW).NPJ Syst Biol Appl. 2017 Sep 19;3:26. doi: 10.1038/s41540-017-0028-x. eCollection 2017. NPJ Syst Biol Appl. 2017. PMID: 28944079 Free PMC article. Review.
-
The Centrosome, a Multitalented Renaissance Organelle.Cold Spring Harb Perspect Biol. 2016 Dec 1;8(12):a025049. doi: 10.1101/cshperspect.a025049. Cold Spring Harb Perspect Biol. 2016. PMID: 27908937 Free PMC article. Review.
-
Cortical Cyclin A controls spindle orientation during asymmetric cell divisions in Drosophila.Nat Commun. 2022 May 17;13(1):2723. doi: 10.1038/s41467-022-30182-1. Nat Commun. 2022. PMID: 35581185 Free PMC article.
-
A guiding torch at the poles: the multiple roles of spindle microtubule-organizing centers during cell division.Cell Cycle. 2020 Jun;19(12):1405-1421. doi: 10.1080/15384101.2020.1754586. Epub 2020 May 13. Cell Cycle. 2020. PMID: 32401610 Free PMC article. Review.
-
BRCA2 mediates centrosome cohesion via an interaction with cytoplasmic dynein.Cell Cycle. 2016 Aug 17;15(16):2145-2156. doi: 10.1080/15384101.2016.1195531. Epub 2016 Jul 19. Cell Cycle. 2016. PMID: 27433848 Free PMC article.
References
-
- Wiese C, Zheng Y. Microtubule nucleation: Gamma-tubulin and beyond. J Cell Sci. 2006;119:4143–4153. - PubMed
-
- Cuschieri L, Nguyen T, Vogel J. Control at the cell center: The role of spindle poles in cytoskeletal organization and cell cycle regulation. Cell Cycle. 2007;6:2788–2794. - PubMed
-
- Doxsey S, McCollum D, Theurkauf W. Centrosomes in cellular regulation. Annu Rev Cell Dev Biol. 2005;21:411–434. - PubMed
-
- Doxsey S, Zimmerman W, Mikule K. Centrosome control of the cell cycle. Trends Cell Biol. 2005;15:303–311. - PubMed
-
- Hinchcliffe EH, Miller FJ, Cham M, Khodjakov A, Sluder G. Requirement of a centrosomal activity for cell cycle progression through G1 into S phase. Science. 2001;291:1547–1550. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

