Molecular keys of the tropism of integration of the cholera toxin phage
- PMID: 20133778
- PMCID: PMC2840090
- DOI: 10.1073/pnas.0910212107
Molecular keys of the tropism of integration of the cholera toxin phage
Abstract
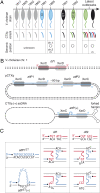
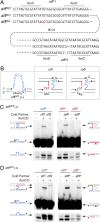
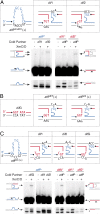
Cholera toxin is encoded in the genome of CTXvarphi, a lysogenic filamentous phage of Vibrio cholerae. CTXvarphi variants contribute to the genetic diversity of cholera epidemic strains. It has been shown that the El Tor variant of CTXvarphi hijacks XerC and XerD, two host-encoded tyrosine recombinases that normally function to resolve chromosome dimers, to integrate at dif1, the dimer resolution site of the larger of the two V. cholerae chromosomes. However, the exact mechanism of integration of CTXvarphi and the rules governing its integration remained puzzling, with phage variants integrated at either or both dimer resolution sites of the two V. cholerae chromosomes. We designed a genetic system to determine experimentally the tropism of integration of CTXvarphi and thus define rules of compatibility between phage variants and dimer resolution sites. We then showed in vitro how these rules are explained by the direct integration of the single-stranded phage genome into the double-stranded bacterial genome. Finally, we showed how the evolution of phage attachment and chromosome dimer resolution sites contributes to the generation of genetic diversity among cholera epidemic strains.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Comment in
-
Efficiency and specificity of CTXphi chromosomal integration: dif makes all the difference.Proc Natl Acad Sci U S A. 2010 Mar 2;107(9):3951-2. doi: 10.1073/pnas.1000310107. Proc Natl Acad Sci U S A. 2010. PMID: 20197438 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

