The circadian output gene takeout is regulated by Pdp1epsilon
- PMID: 20133786
- PMCID: PMC2823862
- DOI: 10.1073/pnas.0906422107
The circadian output gene takeout is regulated by Pdp1epsilon
Abstract
The circadian clock controls many circadian outputs. Although a large number of transcripts are affected by the circadian oscillator, very little is known about their regulation and function. We show here that the Drosophila takeout gene, one of the output genes of the circadian oscillator, is regulated similarly to the circadian clock genes Clock (Clk) and cry. takeout RNA levels are at constant high levels in Clk(JRK) mutants. The circadian transcription factor PAR domain protein 1 (Pdp1epsilon) is a transcription factor that had previously been postulated to control clock output genes, particularly genes regulated similarly to Clk. In agreement with this, we show here that Pdp1epsilon is a regulator of takeout. Takeout levels are low in flies with reduced Pdp1epsilon and high in flies with increased amounts of Pdp1epsilon. Furthermore, flies with reduced or elevated Pdp1epsilon levels in the fat body display courtship defects, identifying Pdp1epsilon as an important transcriptional regulator in that tissue.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
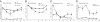
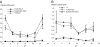

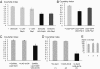
Similar articles
-
An isoform-specific mutant reveals a role of PDP1 epsilon in the circadian oscillator.J Neurosci. 2009 Sep 2;29(35):10920-7. doi: 10.1523/JNEUROSCI.2133-09.2009. J Neurosci. 2009. PMID: 19726650 Free PMC article.
-
PDP1epsilon functions downstream of the circadian oscillator to mediate behavioral rhythms.J Neurosci. 2007 Mar 7;27(10):2539-47. doi: 10.1523/JNEUROSCI.4870-06.2007. J Neurosci. 2007. PMID: 17344391 Free PMC article.
-
Targeted inhibition of Pdp1epsilon abolishes the circadian behavior of Drosophila melanogaster.Biochem Biophys Res Commun. 2007 Dec 14;364(2):294-300. doi: 10.1016/j.bbrc.2007.10.009. Epub 2007 Oct 12. Biochem Biophys Res Commun. 2007. PMID: 17950247
-
Transcriptional feedback loop regulation, function, and ontogeny in Drosophila.Cold Spring Harb Symp Quant Biol. 2007;72:437-44. doi: 10.1101/sqb.2007.72.009. Cold Spring Harb Symp Quant Biol. 2007. PMID: 18419302 Free PMC article. Review.
-
Circadian Rhythms and Sleep in Drosophila melanogaster.Genetics. 2017 Apr;205(4):1373-1397. doi: 10.1534/genetics.115.185157. Genetics. 2017. PMID: 28360128 Free PMC article. Review.
Cited by
-
Molecular genetic analysis of circadian timekeeping in Drosophila.Adv Genet. 2011;74:141-73. doi: 10.1016/B978-0-12-387690-4.00005-2. Adv Genet. 2011. PMID: 21924977 Free PMC article. Review.
-
Non-Pleiotropic Coupling of Daily and Seasonal Temporal Isolation in the European Corn Borer.Genes (Basel). 2018 Mar 26;9(4):180. doi: 10.3390/genes9040180. Genes (Basel). 2018. PMID: 29587435 Free PMC article.
-
Clk post-transcriptional control denoises circadian transcription both temporally and spatially.Nat Commun. 2015 May 8;6:7056. doi: 10.1038/ncomms8056. Nat Commun. 2015. PMID: 25952406 Free PMC article.
-
Evidence that natural selection maintains genetic variation for sleep in Drosophila melanogaster.BMC Evol Biol. 2015 Mar 13;15:41. doi: 10.1186/s12862-015-0316-2. BMC Evol Biol. 2015. PMID: 25887180 Free PMC article.
-
Mated Drosophila melanogaster females consume more amino acids during the dark phase.PLoS One. 2017 Feb 27;12(2):e0172886. doi: 10.1371/journal.pone.0172886. eCollection 2017. PLoS One. 2017. PMID: 28241073 Free PMC article.
References
-
- Hall JC. Genetics and Molecular Biology of Rhythms in Drosophila and Other Insects. Adv Genet. 2003;48:1–280. - PubMed
-
- Hardin PE. The circadian timekeeping system of Drosophila. Curr Biol. 2005;15:R714–R722. - PubMed
-
- Glossop NR, Lyons LC, Hardin PE. Interlocked feedback loops within the Drosophila circadian oscillator. Science. 1999;286:766–768. - PubMed
-
- Darlington TK, et al. Closing the circadian loop: CLOCK-induced transcription of its own inhibitors per and tim. Science. 1998;280:1599–1603. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

