Transcriptional coactivator HCF-1 couples the histone chaperone Asf1b to HSV-1 DNA replication components
- PMID: 20133788
- PMCID: PMC2809754
- DOI: 10.1073/pnas.0911128107
Transcriptional coactivator HCF-1 couples the histone chaperone Asf1b to HSV-1 DNA replication components
Abstract
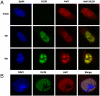
The cellular transcriptional coactivator HCF-1 interacts with numerous transcription factors as well as other coactivators and is a component of multiple chromatin modulation complexes. The protein is essential for the expression of the immediate early genes of both herpes simplex virus (HSV) and varicella zoster virus and functions, in part, by coupling chromatin modification components including the Set1 or MLL1 histone methyltransferases and the histone demethylase LSD1 to promote the installation of positive chromatin marks and the activation of viral immediately early gene transcription. Although studies have investigated the role of HCF-1 in both cellular and viral transcription, little is known about other processes that the protein may be involved in. Here we demonstrate that HCF-1 localizes to sites of HSV replication late in infection. HCF-1 interacts directly and simultaneously with both HSV DNA replication proteins and the cellular histone chaperone Asf1b, a protein that regulates the progression of cellular DNA replication forks via chromatin reorganization. Asf1b localizes with HCF-1 in viral replication foci and depletion of Asf1b results in significantly reduced viral DNA accumulation. The results support a model in which the transcriptional coactivator HCF-1 is a component of the HSV DNA replication assembly and promotes viral DNA replication by coupling Asf1b to DNA replication components. This coupling provides a novel function for HCF-1 and insights into the mechanisms of modulating chromatin during DNA replication.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






Similar articles
-
The coactivator host cell factor-1 mediates Set1 and MLL1 H3K4 trimethylation at herpesvirus immediate early promoters for initiation of infection.Proc Natl Acad Sci U S A. 2007 Jun 26;104(26):10835-40. doi: 10.1073/pnas.0704351104. Epub 2007 Jun 19. Proc Natl Acad Sci U S A. 2007. PMID: 17578910 Free PMC article.
-
Inhibition of the histone demethylase LSD1 blocks alpha-herpesvirus lytic replication and reactivation from latency.Nat Med. 2009 Nov;15(11):1312-7. doi: 10.1038/nm.2051. Epub 2009 Oct 25. Nat Med. 2009. PMID: 19855399 Free PMC article.
-
Combinatorial transcription of herpes simplex virus and varicella zoster virus immediate early genes is strictly determined by the cellular coactivator HCF-1.J Biol Chem. 2005 Jan 14;280(2):1369-75. doi: 10.1074/jbc.M410178200. Epub 2004 Nov 1. J Biol Chem. 2005. PMID: 15522876
-
Control of alpha-herpesvirus IE gene expression by HCF-1 coupled chromatin modification activities.Biochim Biophys Acta. 2010 Mar-Apr;1799(3-4):257-65. doi: 10.1016/j.bbagrm.2009.08.003. Epub 2009 Aug 12. Biochim Biophys Acta. 2010. PMID: 19682612 Free PMC article. Review.
-
The dynamics of HCF-1 modulation of herpes simplex virus chromatin during initiation of infection.Viruses. 2013 May 22;5(5):1272-91. doi: 10.3390/v5051272. Viruses. 2013. PMID: 23698399 Free PMC article. Review.
Cited by
-
Herpes simplex virus 1 DNA is in unstable nucleosomes throughout the lytic infection cycle, and the instability of the nucleosomes is independent of DNA replication.J Virol. 2012 Oct;86(20):11287-300. doi: 10.1128/JVI.01468-12. Epub 2012 Aug 8. J Virol. 2012. PMID: 22875975 Free PMC article.
-
Identification of a Polypeptide Inhibitor of O-GlcNAc Transferase with Picomolar Affinity.J Am Chem Soc. 2024 Sep 25;146(38):26320-26330. doi: 10.1021/jacs.4c08656. Epub 2024 Sep 14. J Am Chem Soc. 2024. PMID: 39276112 Free PMC article.
-
LINC00665 Promotes the Progression of Multiple Myeloma by Adsorbing miR-214-3p and Positively Regulating the Expression of PSMD10 and ASF1B.Onco Targets Ther. 2020 Jul 7;13:6511-6522. doi: 10.2147/OTT.S241627. eCollection 2020. Onco Targets Ther. 2020. Retraction in: Onco Targets Ther. 2021 Jun 01;14:3535. doi: 10.2147/OTT.S321549. PMID: 32764956 Free PMC article. Retracted.
-
Snapshots: chromatin control of viral infection.Virology. 2013 Jan 5;435(1):141-56. doi: 10.1016/j.virol.2012.09.023. Virology. 2013. PMID: 23217624 Free PMC article. Review.
-
The pre-NH(2)-terminal domain of the herpes simplex virus 1 DNA polymerase catalytic subunit is required for efficient viral replication.J Virol. 2012 Oct;86(20):11057-65. doi: 10.1128/JVI.01034-12. Epub 2012 Aug 8. J Virol. 2012. PMID: 22875965 Free PMC article.
References
-
- Kristie TM, Liang Y, Vogel JL. Control of alpha-herpesvirus IE gene expression by HCF-1 coupled chromatin modification activities. Biochim Biophys Acta. 2009 doi: 10.1016/j.bbagrm.2009.08. - DOI - PMC - PubMed
-
- Fujiki R, et al. GlcNAcylation of a histone methyltransferase in retinoic-acid-induced granulopoiesis. Nature. 2009;459:455–459. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

