Genome-minimized Streptomyces host for the heterologous expression of secondary metabolism
- PMID: 20133795
- PMCID: PMC2823899
- DOI: 10.1073/pnas.0914833107
Genome-minimized Streptomyces host for the heterologous expression of secondary metabolism
Abstract
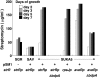
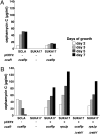
To construct a versatile model host for heterologous expression of genes encoding secondary metabolite biosynthesis, the genome of the industrial microorganism Streptomyces avermitilis was systematically deleted to remove nonessential genes. A region of more than 1.4 Mb was deleted stepwise from the 9.02-Mb S. avermitilis linear chromosome to generate a series of defined deletion mutants, corresponding to 83.12-81.46% of the wild-type chromosome, that did not produce any of the major endogenous secondary metabolites found in the parent strain. The suitability of the mutants as hosts for efficient production of foreign metabolites was shown by heterologous expression of three different exogenous biosynthetic gene clusters encoding the biosynthesis of streptomycin (from S. griseus Institute for Fermentation, Osaka [IFO] 13350), cephamycin C (from S. clavuligerus American type culture collection (ATCC) 27064), and pladienolide (from S. platensis Mer-11107). Both streptomycin and cephamycin C were efficiently produced by individual transformants at levels higher than those of the native-producing species. Although pladienolide was not produced by a deletion mutant transformed with the corresponding intact biosynthetic gene cluster, production of the macrolide was enabled by introduction of an extra copy of the regulatory gene pldR expressed under control of an alternative promoter. Another mutant optimized for terpenoid production efficiently produced the plant terpenoid intermediate, amorpha-4,11-diene, by introduction of a synthetic gene optimized for Streptomyces codon usage. These findings highlight the strength and flexibility of engineered S. avermitilis as a model host for heterologous gene expression, resulting in the production of exogenous natural and unnatural metabolites.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Engineering of a genome-reduced host: practical application of synthetic biology in the overproduction of desired secondary metabolites.Protein Cell. 2010 Jul;1(7):621-6. doi: 10.1007/s13238-010-0073-3. Epub 2010 Jul 29. Protein Cell. 2010. PMID: 21203934 Free PMC article. Review.
-
Engineered Streptomyces avermitilis host for heterologous expression of biosynthetic gene cluster for secondary metabolites.ACS Synth Biol. 2013 Jul 19;2(7):384-96. doi: 10.1021/sb3001003. Epub 2013 Jan 17. ACS Synth Biol. 2013. PMID: 23654282 Free PMC article.
-
Genome mining of the Streptomyces avermitilis genome and development of genome-minimized hosts for heterologous expression of biosynthetic gene clusters.J Ind Microbiol Biotechnol. 2014 Feb;41(2):233-50. doi: 10.1007/s10295-013-1327-x. Epub 2013 Aug 29. J Ind Microbiol Biotechnol. 2014. PMID: 23990133 Review.
-
Rational construction of genome-reduced and high-efficient industrial Streptomyces chassis based on multiple comparative genomic approaches.Microb Cell Fact. 2019 Jan 28;18(1):16. doi: 10.1186/s12934-019-1055-7. Microb Cell Fact. 2019. PMID: 30691531 Free PMC article.
-
Heterologous Expression of Spinosyn Biosynthetic Gene Cluster in Streptomyces Species Is Dependent on the Expression of Rhamnose Biosynthesis Genes.J Mol Microbiol Biotechnol. 2017;27(3):190-198. doi: 10.1159/000477543. Epub 2017 Aug 19. J Mol Microbiol Biotechnol. 2017. PMID: 28848197
Cited by
-
Iron-sulphur protein catalysed [4+2] cycloadditions in natural product biosynthesis.Nat Commun. 2024 Jul 10;15(1):5779. doi: 10.1038/s41467-024-50142-1. Nat Commun. 2024. PMID: 38987535 Free PMC article.
-
Recent advances in natural products exploitation in Streptomyces via synthetic biology.Eng Life Sci. 2019 Mar 12;19(6):452-462. doi: 10.1002/elsc.201800137. eCollection 2019 Jun. Eng Life Sci. 2019. PMID: 32625022 Free PMC article. Review.
-
Minimal polyketide pathway expression in an actinorhodin cluster-deleted and regulation-stimulated Streptomyces coelicolor.J Ind Microbiol Biotechnol. 2012 May;39(5):805-11. doi: 10.1007/s10295-011-1083-8. Epub 2012 Jan 18. J Ind Microbiol Biotechnol. 2012. PMID: 22252445
-
Heterologous production of cyanobacterial compounds.J Ind Microbiol Biotechnol. 2021 Jun 4;48(3-4):kuab003. doi: 10.1093/jimb/kuab003. J Ind Microbiol Biotechnol. 2021. PMID: 33928376 Free PMC article. Review.
-
Heterologous expression of 8-demethyl-tetracenomycin (8-dmtc) affected Streptomyces coelicolor life cycle.Braz J Microbiol. 2021 Sep;52(3):1107-1118. doi: 10.1007/s42770-021-00499-y. Epub 2021 Apr 20. Braz J Microbiol. 2021. PMID: 33876406 Free PMC article.
References
-
- Ikeda H, et al. Complete genome sequence and comparative analysis of the industrial microorganism Streptomyces avermitilis . Nat Biotechnol. 2003;21:526–531. - PubMed
-
- Bentley SD, et al. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2) Nature. 2002;417:141–147. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

