Alternative end-joining catalyzes robust IgH locus deletions and translocations in the combined absence of ligase 4 and Ku70
- PMID: 20133803
- PMCID: PMC2840344
- DOI: 10.1073/pnas.0915067107
Alternative end-joining catalyzes robust IgH locus deletions and translocations in the combined absence of ligase 4 and Ku70
Erratum in
- Proc Natl Acad Sci U S A. 2013 Apr 2;110(14):5731
Abstract
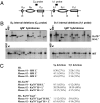
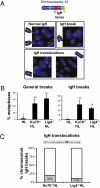
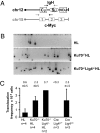
Class switch recombination (CSR) in B lymphocytes is initiated by introduction of multiple DNA double-strand breaks (DSBs) into switch (S) regions that flank immunoglobulin heavy chain (IgH) constant region exons. CSR is completed by joining a DSB in the donor S mu to a DSB in a downstream acceptor S region (e.g., S gamma1) by end-joining. In normal cells, many CSR junctions are mediated by classical nonhomologous end-joining (C-NHEJ), which employs the Ku70/80 complex for DSB recognition and XRCC4/DNA ligase 4 for ligation. Alternative end-joining (A-EJ) mediates CSR, at reduced levels, in the absence of C-NHEJ, even in combined absence of Ku70 and ligase 4, demonstrating an A-EJ pathway totally distinct from C-NHEJ. Multiple DSBs are introduced into S mu during CSR, with some being rejoined or joined to each other to generate internal switch deletions (ISDs). In addition, S-region DSBs can be joined to other chromosomes to generate translocations, the level of which is increased by absence of a single C-NHEJ component (e.g., XRCC4). We asked whether ISD and S-region translocations occur in the complete absence of C-NHEJ (e.g., in Ku70/ligase 4 double-deficient B cells). We found, unexpectedly, that B-cell activation for CSR generates substantial ISD in both S mu and S gamma1 and that ISD in both is greatly increased by the absence of C-NHEJ. IgH chromosomal translocations to the c-myc oncogene also are augmented in the combined absence of Ku70 and ligase 4. We discuss the implications of these findings for A-EJ in normal and abnormal DSB repair.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Honjo T, Kinoshita K, Muramatsu M. Molecular mechanism of class switch recombination: Linkage with somatic hypermutation. Annu Rev Immunol. 2002;20:165–196. - PubMed
-
- Chaudhuri J, et al. Evolution of the immunoglobulin heavy chain class switch recombination mechanism. Adv Immunol. 2007;94:157–214. - PubMed
-
- Muramatsu M, et al. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. - PubMed
-
- Catalan N, et al. The block in immunoglobulin class switch recombination caused by activation-induced cytidine deaminase deficiency occurs prior to the generation of DNA double strand breaks in switch mu region. J Immunol. 2003;171:2504–2509. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI037526/AI/NIAID NIH HHS/United States
- T32CA09382-26/CA/NCI NIH HHS/United States
- U19 AI031541/AI/NIAID NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- P01 CA092625/CA/NCI NIH HHS/United States
- AI077595/AI/NIAID NIH HHS/United States
- T32 CA009382/CA/NCI NIH HHS/United States
- P01 AI031541/AI/NIAID NIH HHS/United States
- R01 AI077595/AI/NIAID NIH HHS/United States
- R37 AI077595/AI/NIAID NIH HHS/United States
- CA092625/CA/NCI NIH HHS/United States
- AI037526/AI/NIAID NIH HHS/United States
- R37 AI037526/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

