PAS domain containing chemoreceptor couples dynamic changes in metabolism with chemotaxis
- PMID: 20133866
- PMCID: PMC2836669
- DOI: 10.1073/pnas.0910055107
PAS domain containing chemoreceptor couples dynamic changes in metabolism with chemotaxis
Abstract
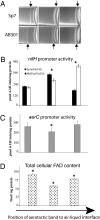
Chemoreceptors provide sensory specificity and sensitivity that enable motile bacteria to seek optimal positions for growth and metabolism in gradients of various physicochemical cues. Despite the abundance of chemoreceptors, little is known regarding the sensory specificity and the exact contribution of individual chemoreceptors to the lifestyle of bacteria. Azospirillum brasilense are motile bacteria that can fix atmospheric nitrogen under microaerophilic conditions. Here, we characterized a chemoreceptor in this organism, named AerC, which functions as a redox sensor that enables the cells to seek microaerophilic conditions that support optimum nitrogen fixation. AerC is a representative of a widespread class of soluble chemoreceptors that monitor changes in the redox status of the electron transport system via the FAD cofactor associated with its PAS domains. In A. brasilense, AerC clusters at the cell poles. Its cellular localization and contribution to the behavioral response correlate with its expression pattern and with changes in the overall cellular FAD content under nitrogen-fixing conditions. AerC-mediated energy taxis in A. brasilense prevails under conditions of nitrogen fixation, illustrating a strategy by which cells optimize chemosensing to signaling cues that directly affect current metabolic activities and thus revealing a mechanism by which chemotaxis is coordinated with dynamic changes in cell physiology.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Azospirillum brasilense AerC and Tlp4b Cytoplasmic Chemoreceptors Are Promiscuous and Interact with the Two Membrane-Bound Chemotaxis Signaling Clusters Mediating Chemotaxis Responses.J Bacteriol. 2023 Jun 27;205(6):e0048422. doi: 10.1128/jb.00484-22. Epub 2023 May 31. J Bacteriol. 2023. PMID: 37255486 Free PMC article.
-
An Azospirillum brasilense chemoreceptor that mediates nitrate chemotaxis has conditional roles in the colonization of plant roots.Appl Environ Microbiol. 2024 Jun 18;90(6):e0076024. doi: 10.1128/aem.00760-24. Epub 2024 May 22. Appl Environ Microbiol. 2024. PMID: 38775579 Free PMC article.
-
An energy taxis transducer promotes root colonization by Azospirillum brasilense.J Bacteriol. 2004 Oct;186(19):6595-604. doi: 10.1128/JB.186.19.6595-6604.2004. J Bacteriol. 2004. PMID: 15375141 Free PMC article.
-
Aerotaxis and other energy-sensing behavior in bacteria.Annu Rev Microbiol. 1999;53:103-28. doi: 10.1146/annurev.micro.53.1.103. Annu Rev Microbiol. 1999. PMID: 10547687 Review.
-
Regulation of nitrogen fixation in Azospirillum brasilense.FEMS Microbiol Lett. 1997 Jul 15;152(2):195-204. doi: 10.1111/j.1574-6968.1997.tb10428.x. FEMS Microbiol Lett. 1997. PMID: 9231412 Review.
Cited by
-
Chemotaxis Control of Transient Cell Aggregation.J Bacteriol. 2015 Oct;197(20):3230-7. doi: 10.1128/JB.00121-15. Epub 2015 Jul 27. J Bacteriol. 2015. PMID: 26216846 Free PMC article. Review.
-
A Chemotaxis Receptor Modulates Nodulation during the Azorhizobium caulinodans-Sesbania rostrata Symbiosis.Appl Environ Microbiol. 2016 May 16;82(11):3174-84. doi: 10.1128/AEM.00230-16. Print 2016 Jun 1. Appl Environ Microbiol. 2016. PMID: 26994081 Free PMC article.
-
Azospirillum brasilense AerC and Tlp4b Cytoplasmic Chemoreceptors Are Promiscuous and Interact with the Two Membrane-Bound Chemotaxis Signaling Clusters Mediating Chemotaxis Responses.J Bacteriol. 2023 Jun 27;205(6):e0048422. doi: 10.1128/jb.00484-22. Epub 2023 May 31. J Bacteriol. 2023. PMID: 37255486 Free PMC article.
-
An Azospirillum brasilense chemoreceptor that mediates nitrate chemotaxis has conditional roles in the colonization of plant roots.Appl Environ Microbiol. 2024 Jun 18;90(6):e0076024. doi: 10.1128/aem.00760-24. Epub 2024 May 22. Appl Environ Microbiol. 2024. PMID: 38775579 Free PMC article.
-
Tools for genetic manipulation of the plant growth-promoting bacterium Azospirillum amazonense.BMC Microbiol. 2011 May 16;11:107. doi: 10.1186/1471-2180-11-107. BMC Microbiol. 2011. PMID: 21575234 Free PMC article.
References
-
- Wadhams GH, Armitage JP. Making sense of it all: Bacterial chemotaxis. Nat Rev Mol Cell Biol. 2004;5:1024–1037. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

