African great apes are natural hosts of multiple related malaria species, including Plasmodium falciparum
- PMID: 20133889
- PMCID: PMC2824423
- DOI: 10.1073/pnas.0914440107
African great apes are natural hosts of multiple related malaria species, including Plasmodium falciparum
Abstract
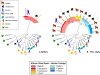
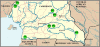
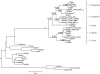
Plasmodium reichenowi, a chimpanzee parasite, was until very recently the only known close relative of Plasmodium falciparum, the most virulent agent of human malaria. Recently, Plasmodium gaboni, another closely related chimpanzee parasite, was discovered, suggesting that the diversity of Plasmodium circulating in great apes in Africa might have been underestimated. It was also recently shown that P. reichenowi is a geographically widespread and genetically diverse chimpanzee parasite and that the world diversity of P. falciparum is fully included within the much broader genetic diversity of P. reichenowi. The evidence indicates that all extant populations of P. falciparum originated from P. reichenowi, likely by a single transfer from chimpanzees. In this work, we have studied the diversity of Plasmodium species infecting chimpanzees and gorillas in Central Africa (Cameroon and Gabon) from both wild-living and captive animals. The studies in wild apes used noninvasive sampling methods. We confirm the presence of P. reichenowi and P. gaboni in wild chimpanzees. Moreover, our results reveal the existence of an unexpected genetic diversity of Plasmodium lineages circulating in gorillas. We show that gorillas are naturally infected by two related lineages of parasites that have not been described previously, herein referred to as Plasmodium GorA and P. GorB, but also by P. falciparum, a species previously considered as strictly human specific. The continuously increasing contacts between humans and primate populations raise concerns about further reciprocal host transfers of these pathogens.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Escalante AA, Barrio E, Ayala FJ. Evolutionary origin of human and primate malarias: Evidence from the circumsporozoite protein gene. Mol Biol Evol. 1995;12:616–626. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

