Regulation of CD1 antigen-presenting complex stability
- PMID: 20133943
- PMCID: PMC2852931
- DOI: 10.1074/jbc.M109.077933
Regulation of CD1 antigen-presenting complex stability
Abstract
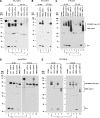
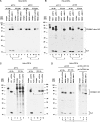
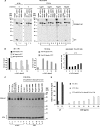
For major histocompatibility complex class I and II molecules, the binding of specific peptide antigens is essential for assembly and trafficking and is at the center of their quality control mechanism. However, the role of lipid antigen binding in stabilization and quality control of CD1 heavy chain (HC).beta(2)-microglobulin (beta(2)m) complexes is unclear. Furthermore, the distinct trafficking and loading routes of CD1 proteins take them from mildly acidic pH in early endososmal compartments (pH 6.0) to markedly acidic pH in lysosomes (pH 5.0) and back to neutral pH of the cell surface (pH 7.4). Here, we present evidence that the stability of each CD1 HC.beta(2)m complex is determined by the distinct pH optima identical to that of the intracellular compartments in which each CD1 isoform resides. Although stable at acidic endosomal pH, complexes are only stable at cell surface pH 7.4 when bound to specific lipid antigens. The proposed model outlines a quality control program that allows lipid exchange at low endosomal pH without dissociation of the CD1 HC.beta(2)m complex and then stabilizes the antigen-loaded complex at neutral pH at the cell surface.
Figures




Similar articles
-
Cytoplasmic tail-dependent localization of CD1b antigen-presenting molecules to MIICs.Science. 1996 Jul 19;273(5273):349-52. doi: 10.1126/science.273.5273.349. Science. 1996. PMID: 8662520
-
Activation of human CD4+ T cells by targeting MHC class II epitopes to endosomal compartments using human CD1 tail sequences.Immunology. 2007 Dec;122(4):522-31. doi: 10.1111/j.1365-2567.2007.02666.x. Epub 2007 Jul 16. Immunology. 2007. PMID: 17635609 Free PMC article.
-
Four pathways of CD1 antigen presentation to T cells.Curr Opin Immunol. 2017 Jun;46:127-133. doi: 10.1016/j.coi.2017.07.013. Epub 2017 Jul 28. Curr Opin Immunol. 2017. PMID: 28756303 Free PMC article. Review.
-
The Third Way: Progress on pathways of antigen processing and presentation by CD1.Immunol Cell Biol. 2004 Jun;82(3):295-306. doi: 10.1111/j.0818-9641.2004.01258.x. Immunol Cell Biol. 2004. PMID: 15186261 Review.
-
CD1d endosomal trafficking is independently regulated by an intrinsic CD1d-encoded tyrosine motif and by the invariant chain.Immunity. 2001 Dec;15(6):897-908. doi: 10.1016/s1074-7613(01)00240-0. Immunity. 2001. PMID: 11754812
Cited by
-
Role of Group 1 CD1-Restricted T Cells in Infectious Disease.Front Immunol. 2015 Jun 29;6:337. doi: 10.3389/fimmu.2015.00337. eCollection 2015. Front Immunol. 2015. PMID: 26175733 Free PMC article. Review.
-
Endogenous lipid antigens for invariant natural killer T cells hold the reins in adipose tissue homeostasis.Immunology. 2018 Feb;153(2):179-189. doi: 10.1111/imm.12839. Epub 2017 Oct 26. Immunology. 2018. PMID: 28898395 Free PMC article. Review.
-
Saposins modulate human invariant Natural Killer T cells self-reactivity and facilitate lipid exchange with CD1d molecules during antigen presentation.Proc Natl Acad Sci U S A. 2013 Dec 3;110(49):E4753-61. doi: 10.1073/pnas.1310050110. Epub 2013 Nov 18. Proc Natl Acad Sci U S A. 2013. PMID: 24248359 Free PMC article.
-
Analyzing antigen recognition by Natural Killer T cells.Methods Mol Biol. 2013;960:557-572. doi: 10.1007/978-1-62703-218-6_41. Methods Mol Biol. 2013. PMID: 23329514 Free PMC article.
-
The CD1 size problem: lipid antigens, ligands, and scaffolds.Cell Mol Life Sci. 2014 Aug;71(16):3069-79. doi: 10.1007/s00018-014-1603-6. Cell Mol Life Sci. 2014. PMID: 24658584 Free PMC article. Review.
References
-
- Cohen N. R., Garg S., Brenner M. B. (2009) Adv. Immunol. 102, 1–94 - PubMed
-
- Kang S. J., Cresswell P. (2002) J. Biol. Chem. 277, 44838–44844 - PubMed
-
- De Silva A. D., Park J. J., Matsuki N., Stanic A. K., Brutkiewicz R. R., Medof M. E., Joyce S. (2002) J. Immunol. 168, 723–733 - PubMed
-
- Sugita M., Grant E. P., van Donselaar E., Hsu V. W., Rogers R. A., Peters P. J., Brenner M. B. (1999) Immunity 11, 743–752 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

