Prediction of human functional genetic networks from heterogeneous data using RVM-based ensemble learning
- PMID: 20134029
- PMCID: PMC2832827
- DOI: 10.1093/bioinformatics/btq044
Prediction of human functional genetic networks from heterogeneous data using RVM-based ensemble learning
Abstract
Motivation: Three major problems confront the construction of a human genetic network from heterogeneous genomics data using kernel-based approaches: definition of a robust gold-standard negative set, large-scale learning and massive missing data values.
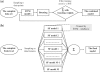
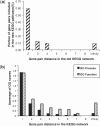
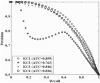
Results: The proposed graph-based approach generates a robust GSN for the training process of genetic network construction. The RVM-based ensemble model that combines AdaBoost and reduced-feature yields improved performance on large-scale learning problems with massive missing values in comparison to Naïve Bayes.
Contact: dargenio@bmsr.usc.edu
Supplementary information: Supplementary material is available at Bioinformatics online.
Figures
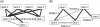
Similar articles
-
forgeNet: a graph deep neural network model using tree-based ensemble classifiers for feature graph construction.Bioinformatics. 2020 Jun 1;36(11):3507-3515. doi: 10.1093/bioinformatics/btaa164. Bioinformatics. 2020. PMID: 32163118 Free PMC article.
-
MetaKTSP: a meta-analytic top scoring pair method for robust cross-study validation of omics prediction analysis.Bioinformatics. 2016 Jul 1;32(13):1966-73. doi: 10.1093/bioinformatics/btw115. Epub 2016 Mar 2. Bioinformatics. 2016. PMID: 27153719 Free PMC article.
-
mRMRe: an R package for parallelized mRMR ensemble feature selection.Bioinformatics. 2013 Sep 15;29(18):2365-8. doi: 10.1093/bioinformatics/btt383. Epub 2013 Jul 3. Bioinformatics. 2013. PMID: 23825369
-
What is bioinformatics? A proposed definition and overview of the field.Methods Inf Med. 2001;40(4):346-58. Methods Inf Med. 2001. PMID: 11552348 Review.
-
Promoting synergistic research and education in genomics and bioinformatics.BMC Genomics. 2008;9 Suppl 1(Suppl 1):I1. doi: 10.1186/1471-2164-9-S1-I1. BMC Genomics. 2008. PMID: 18366597 Free PMC article. Review.
Cited by
-
A comparison of graph- and kernel-based -omics data integration algorithms for classifying complex traits.BMC Bioinformatics. 2017 Dec 6;18(1):539. doi: 10.1186/s12859-017-1982-4. BMC Bioinformatics. 2017. PMID: 29212468 Free PMC article.
-
MoNETA: MultiOmics Network Embedding for SubType Analysis.NAR Genom Bioinform. 2024 Oct 16;6(4):lqae141. doi: 10.1093/nargab/lqae141. eCollection 2024 Sep. NAR Genom Bioinform. 2024. PMID: 39416887 Free PMC article.
-
TARGETgene: a tool for identification of potential therapeutic targets in cancer.PLoS One. 2012;7(8):e43305. doi: 10.1371/journal.pone.0043305. Epub 2012 Aug 31. PLoS One. 2012. PMID: 22952662 Free PMC article.
-
Identification of cancer fusion drivers using network fusion centrality.Bioinformatics. 2013 May 1;29(9):1174-81. doi: 10.1093/bioinformatics/btt131. Epub 2013 Mar 16. Bioinformatics. 2013. PMID: 23505294 Free PMC article.
-
Machine learning: its challenges and opportunities in plant system biology.Appl Microbiol Biotechnol. 2022 May;106(9-10):3507-3530. doi: 10.1007/s00253-022-11963-6. Epub 2022 May 16. Appl Microbiol Biotechnol. 2022. PMID: 35575915 Review.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

