Transcriptional response of Burkholderia cenocepacia J2315 sessile cells to treatments with high doses of hydrogen peroxide and sodium hypochlorite
- PMID: 20137066
- PMCID: PMC2830190
- DOI: 10.1186/1471-2164-11-90
Transcriptional response of Burkholderia cenocepacia J2315 sessile cells to treatments with high doses of hydrogen peroxide and sodium hypochlorite
Abstract
Background: Burkholderia cepacia complex bacteria are opportunistic pathogens, which can cause severe respiratory tract infections in patients with cystic fibrosis (CF). As treatment of infected CF patients is problematic, multiple preventive measures are taken to reduce the infection risk. Besides a stringent segregation policy to prevent patient-to-patient transmission, clinicians also advise patients to clean and disinfect their respiratory equipment on a regular basis. However, problems regarding the efficacy of several disinfection procedures for the removal and/or killing of B. cepacia complex bacteria have been reported. In order to unravel the molecular mechanisms involved in the resistance of biofilm-grown Burkholderia cenocepacia cells against high concentrations of reactive oxygen species (ROS), the present study focussed on the transcriptional response in sessile B. cenocepacia J2315 cells following exposure to high levels of H2O2 or NaOCl.
Results: The exposure to H2O2 and NaOCl resulted in an upregulation of the transcription of 315 (4.4%) and 386 (5.4%) genes, respectively. Transcription of 185 (2.6%) and 331 (4.6%) genes was decreased in response to the respective treatments. Many of the upregulated genes in the NaOCl- and H2O2-treated biofilms are involved in oxidative stress as well as general stress response, emphasizing the importance of the efficient neutralization and scavenging of ROS. In addition, multiple upregulated genes encode proteins that are necessary to repair ROS-induced cellular damage. Unexpectedly, a prolonged treatment with H2O2 also resulted in an increased transcription of multiple phage-related genes. A closer inspection of hybridisation signals obtained with probes targeting intergenic regions led to the identification of a putative 6S RNA.
Conclusion: Our results reveal that the transcription of a large fraction of B. cenocepacia J2315 genes is altered upon exposure of sessile cells to ROS. These observations have highlighted that B. cenocepacia may alter several pathways in response to exposure to ROS and they have led to the identification of many genes not previously implicated in the stress response of this pathogen.
Figures





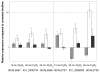

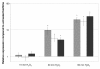
Similar articles
-
Evaluation of the efficacy of disinfection procedures against Burkholderia cenocepacia biofilms.J Hosp Infect. 2008 Dec;70(4):361-8. doi: 10.1016/j.jhin.2008.08.015. Epub 2008 Nov 1. J Hosp Infect. 2008. PMID: 18977555
-
Gene expression changes linked to antimicrobial resistance, oxidative stress, iron depletion and retained motility are observed when Burkholderia cenocepacia grows in cystic fibrosis sputum.BMC Infect Dis. 2008 Sep 19;8:121. doi: 10.1186/1471-2334-8-121. BMC Infect Dis. 2008. PMID: 18801206 Free PMC article.
-
Molecular mechanisms of chlorhexidine tolerance in Burkholderia cenocepacia biofilms.Antimicrob Agents Chemother. 2011 May;55(5):1912-9. doi: 10.1128/AAC.01571-10. Epub 2011 Feb 28. Antimicrob Agents Chemother. 2011. PMID: 21357299 Free PMC article.
-
Burkholderia cenocepacia in cystic fibrosis: epidemiology and molecular mechanisms of virulence.Clin Microbiol Infect. 2010 Jul;16(7):821-30. doi: 10.1111/j.1469-0691.2010.03237.x. Clin Microbiol Infect. 2010. PMID: 20880411 Review.
-
Social interactions in the Burkholderia cepacia complex: biofilms and quorum sensing.Future Microbiol. 2010 Jul;5(7):1087-99. doi: 10.2217/fmb.10.68. Future Microbiol. 2010. PMID: 20632807 Review.
Cited by
-
Cell Wall Recycling-Linked Coregulation of AmpC and PenB β-Lactamases through ampD Mutations in Burkholderia cenocepacia.Antimicrob Agents Chemother. 2015 Dec;59(12):7602-10. doi: 10.1128/AAC.01068-15. Epub 2015 Sep 28. Antimicrob Agents Chemother. 2015. PMID: 26416862 Free PMC article.
-
Bacterial responses to reactive chlorine species.Annu Rev Microbiol. 2013;67:141-60. doi: 10.1146/annurev-micro-102912-142520. Epub 2013 Jun 14. Annu Rev Microbiol. 2013. PMID: 23768204 Free PMC article. Review.
-
Spontaneous and evolutionary changes in the antibiotic resistance of Burkholderia cenocepacia observed by global gene expression analysis.BMC Genomics. 2011 Jul 22;12:373. doi: 10.1186/1471-2164-12-373. BMC Genomics. 2011. PMID: 21781329 Free PMC article.
-
Genetic Determinants Associated With in Vivo Survival of Burkholderia cenocepacia in the Caenorhabditis elegans Model.Front Microbiol. 2018 May 29;9:1118. doi: 10.3389/fmicb.2018.01118. eCollection 2018. Front Microbiol. 2018. PMID: 29896180 Free PMC article.
-
Characterization of 6S RNA in the Lyme disease spirochete.Mol Microbiol. 2020 Feb;113(2):399-417. doi: 10.1111/mmi.14427. Epub 2019 Dec 11. Mol Microbiol. 2020. PMID: 31742773 Free PMC article.
References
-
- Vanlaere E, Baldwin A, Gevers D, Henry D, De Brandt E, LiPuma JJ, Mahenthiralingam E, Speert DP, Dowson C, Vandamme P. Taxon K, a complex within the Burkholderia cepacia complex, comprises at least two novel species, Burkholderia contaminans sp. nov. and Burkholderia lata sp. nov. Int J Syst Evol Microbiol. 2009;59:102–111. doi: 10.1099/ijs.0.001123-0. - DOI - PubMed
-
- Burkholder WH. Sour skin, a bacterial rot of onion bulbs. Phytopathology. 1950;40:115–117.
-
- Burns JL. In: Burkholderia: Molecular Microbiology and Genomics. Coenye T, Vandamme P, editor. Norfolk: Horizon Bioscience; 2007. Antibiotic resistance of Burkholderia spp; pp. 81–91.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

