Complete genome sequence and lifestyle of black-pigmented Corynebacterium aurimucosum ATCC 700975 (formerly C. nigricans CN-1) isolated from a vaginal swab of a woman with spontaneous abortion
- PMID: 20137072
- PMCID: PMC2830990
- DOI: 10.1186/1471-2164-11-91
Complete genome sequence and lifestyle of black-pigmented Corynebacterium aurimucosum ATCC 700975 (formerly C. nigricans CN-1) isolated from a vaginal swab of a woman with spontaneous abortion
Abstract
Background: Corynebacterium aurimucosum is a slightly yellowish, non-lipophilic, facultative anaerobic member of the genus Corynebacterium and predominantly isolated from human clinical specimens. Unusual black-pigmented variants of C. aurimucosum (originally named as C. nigricans) continue to be recovered from the female urogenital tract and they are associated with complications during pregnancy. C. aurimucosum ATCC 700975 (C. nigricans CN-1) was originally isolated from a vaginal swab of a 34-year-old woman who experienced a spontaneous abortion during month six of pregnancy. For a better understanding of the physiology and lifestyle of this potential urogenital pathogen, the complete genome sequence of C. aurimucosum ATCC 700975 was determined.
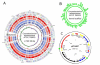
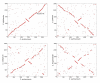
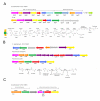
Results: Sequencing and assembly of the C. aurimucosum ATCC 700975 genome yielded a circular chromosome of 2,790,189 bp in size and the 29,037-bp plasmid pET44827. Specific gene sets associated with the central metabolism of C. aurimucosum apparently provide enhanced metabolic flexibility and adaptability in aerobic, anaerobic and low-pH environments, including gene clusters for the uptake and degradation of aromatic amines, L-histidine and L-tartrate as well as a gene region for the formation of selenocysteine and its incorporation into formate dehydrogenase. Plasmid pET44827 codes for a non-ribosomal peptide synthetase that plays the pivotal role in the synthesis of the characteristic black pigment of C. aurimucosum ATCC 700975.
Conclusions: The data obtained by the genome project suggest that C. aurimucosum could be both a resident of the human gut and possibly a pathogen in the female genital tract causing complications during pregnancy. Since hitherto all black-pigmented C. aurimucosum strains have been recovered from female genital source, biosynthesis of the pigment is apparently required for colonization by protecting the bacterial cells against the high hydrogen peroxide concentration in the vaginal environment. The location of the corresponding genes on plasmid pET44827 explains why black-pigmented (formerly C. nigricans) and non-pigmented C. aurimucosum strains were isolated from clinical specimens.
Figures
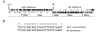


Similar articles
-
Identification of some charcoal-black-pigmented CDC fermentative coryneform group 4 isolates as Rothia dentocariosa and some as Corynebacterium aurimucosum: proposal of Rothia dentocariosa emend. Georg and Brown 1967, Corynebacterium aurimucosum emend. Yassin et al. 2002, and Corynebacterium nigricans Shukla et al. 2003 pro synon. Corynebacterium aurimucosum.J Clin Microbiol. 2004 Sep;42(9):4189-98. doi: 10.1128/JCM.42.9.4189-4198.2004. J Clin Microbiol. 2004. PMID: 15365010 Free PMC article.
-
Corynebacterium nigricans sp. nov.: proposed name for a black-pigmented Corynebacterium species recovered from the human female urogenital tract.J Clin Microbiol. 2003 Sep;41(9):4353-8. doi: 10.1128/JCM.41.9.4353-4358.2003. J Clin Microbiol. 2003. PMID: 12958268 Free PMC article.
-
Isolation and characterization of a black-pigmented Corynebacterium sp. from a woman with spontaneous abortion.J Clin Microbiol. 2001 Mar;39(3):1109-13. doi: 10.1128/JCM.39.3.1109-1113.2001. J Clin Microbiol. 2001. PMID: 11230435 Free PMC article.
-
Is a black-pigmented Corynebacterium species an opportunistic pathogen during pregnancy? Literature review and report of 3 new cases.Clin Infect Dis. 2003 Sep 15;37(6):834-7. doi: 10.1086/377233. Epub 2003 Aug 27. Clin Infect Dis. 2003. PMID: 12955646 Review.
-
Microbiological and clinical features of Corynebacterium urealyticum: urinary tract stones and genomics as the Rosetta Stone.Clin Microbiol Infect. 2008 Jul;14(7):632-43. doi: 10.1111/j.1469-0691.2008.02023.x. Clin Microbiol Infect. 2008. PMID: 18558935 Review.
Cited by
-
Multilocus sequence typing of Corynebacterium ulcerans provides evidence for zoonotic transmission and for increased prevalence of certain sequence types among toxigenic strains.J Clin Microbiol. 2014 Dec;52(12):4318-24. doi: 10.1128/JCM.02291-14. Epub 2014 Oct 15. J Clin Microbiol. 2014. PMID: 25320226 Free PMC article.
-
Clinical Significance of Commensal Gram-Positive Rods Routinely Isolated from Patient Samples.J Clin Microbiol. 2016 Dec;54(12):2928-2936. doi: 10.1128/JCM.01393-16. Epub 2016 Sep 14. J Clin Microbiol. 2016. PMID: 27629905 Free PMC article.
-
Unmasking the invisible enemy: A case report of metagenomics-guided diagnosis and treatment of neonatal septic meningitis caused by Corynebacterium aurimucosum in a preterm infant with neonatal lupus erythematosus.Medicine (Baltimore). 2024 Feb 16;103(7):e35968. doi: 10.1097/MD.0000000000035968. Medicine (Baltimore). 2024. PMID: 38363904 Free PMC article.
-
Recombineering in Corynebacterium glutamicum combined with optical nanosensors: a general strategy for fast producer strain generation.Nucleic Acids Res. 2013 Jul;41(12):6360-9. doi: 10.1093/nar/gkt312. Epub 2013 Apr 28. Nucleic Acids Res. 2013. PMID: 23630315 Free PMC article.
-
The percentage of bacterial genes on leading versus lagging strands is influenced by multiple balancing forces.Nucleic Acids Res. 2012 Sep 1;40(17):8210-8. doi: 10.1093/nar/gks605. Epub 2012 Jun 26. Nucleic Acids Res. 2012. PMID: 22735706 Free PMC article.
References
-
- Goldstein EJ, Citron DM, Merriam CV, Warren YA, Tyrrell KL, Fernandez HT. In vitro activities of doripenem and six comparator drugs against 423 aerobic and anaerobic bacterial isolates from infected diabetic foot wounds. Antimicrob Agents Chemother. 2008;52(2):761–766. doi: 10.1128/AAC.01128-07. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

