Genome-wide investigation and functional characterization of the beta-ketoadipate pathway in the nitrogen-fixing and root-associated bacterium Pseudomonas stutzeri A1501
- PMID: 20137101
- PMCID: PMC2907835
- DOI: 10.1186/1471-2180-10-36
Genome-wide investigation and functional characterization of the beta-ketoadipate pathway in the nitrogen-fixing and root-associated bacterium Pseudomonas stutzeri A1501
Abstract
Background: Soil microorganisms are mainly responsible for the complete mineralization of aromatic compounds that usually originate from plant products or environmental pollutants. In many cases, structurally diverse aromatic compounds can be converted to a small number of structurally simpler intermediates, which are metabolized to tricarboxylic acid intermediates via the beta-ketoadipate pathway. This strategy provides great metabolic flexibility and contributes to increased adaptation of bacteria to their environment. However, little is known about the evolution and regulation of the beta-ketoadipate pathway in root-associated diazotrophs.
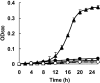
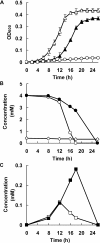
Results: In this report, we performed a genome-wide analysis of the benzoate and 4-hydroxybenzoate catabolic pathways of Pseudomonas stutzeri A1501, with a focus on the functional characterization of the beta-ketoadipate pathway. The P. stutzeri A1501 genome contains sets of catabolic genes involved in the peripheral pathways for catabolism of benzoate (ben) and 4-hydroxybenzoate (pob), and in the catechol (cat) and protocatechuate (pca) branches of the beta-ketoadipate pathway. A particular feature of the catabolic gene organization in A1501 is the absence of the catR and pcaK genes encoding a LysR family regulator and 4-hydroxybenzoate permease, respectively. Furthermore, the BenR protein functions as a transcriptional activator of the ben operon, while transcription from the catBC promoter can be activated in response to benzoate. Benzoate degradation is subject to carbon catabolite repression induced by glucose and acetate in A1501. The HPLC analysis of intracellular metabolites indicated that low concentrations of 4-hydroxybenzoate significantly enhance the ability of A1501 to degrade benzoate.
Conclusions: The expression of genes encoding proteins involved in the beta-ketoadipate pathway is tightly modulated by both pathway-specific and catabolite repression controls in A1501. This strain provides an ideal model system for further study of the evolution and regulation of aromatic catabolic pathways.
Figures






References
-
- Barbe V, Vallenet D, Fonknechten N, Kreimeyer A, Oztas S, Labarre L, Cruveiller S, Robert C, Duprat S, Wincker P, Ornston LN, Weissenbach J, Marlière P, Cohen GN, Médigue C. Unique features revealed by the genome sequence of Acinetobacter sp. ADP1, a versatile and naturally transformation competent bacterium. Nucleic Acids Res. 2004;32(19):5766–5779. doi: 10.1093/nar/gkh910. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

