Homozygosity mapping reveals mutations of GRXCR1 as a cause of autosomal-recessive nonsyndromic hearing impairment
- PMID: 20137778
- PMCID: PMC2820176
- DOI: 10.1016/j.ajhg.2009.12.017
Homozygosity mapping reveals mutations of GRXCR1 as a cause of autosomal-recessive nonsyndromic hearing impairment
Abstract
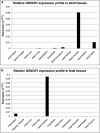
We identified overlapping homozygous regions within the DFNB25 locus in two Dutch and ten Pakistani families with sensorineural autosomal-recessive nonsyndromic hearing impairment (arNSHI). Only one of the families, W98-053, was not consanguineous, and its sibship pointed toward a reduced critical region of 0.9 Mb. This region contained the GRXCR1 gene, and the orthologous mouse gene was described to be mutated in the pirouette (pi) mutant with resulting hearing loss and circling behavior. Sequence analysis of the GRXCR1 gene in hearing-impaired family members revealed splice-site mutations in two Dutch families and a missense and nonsense mutation, respectively, in two Pakistani families. The splice-site mutations are predicted to cause frameshifts and premature stop codons. In family W98-053, this could be confirmed by cDNA analysis. GRXCR1 is predicted to contain a GRX-like domain. GRX domains are involved in reversible S-glutathionylation of proteins and thereby in the modulation of activity and/or localization of these proteins. The missense mutation is located in this domain, whereas the nonsense and splice-site mutations may result in complete or partial absence of the GRX-like domain or of the complete protein. Hearing loss in patients with GRXCR1 mutations is congenital and is moderate to profound. Progression of the hearing loss was observed in family W98-053. Vestibular dysfunction was observed in some but not all affected individuals. Quantitative analysis of GRXCR1 transcripts in fetal and adult human tissues revealed a preferential expression of the gene in fetal cochlea, which may explain the nonsyndromic nature of the hearing impairment.
Copyright (c) 2010 The American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Figures


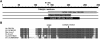


References
-
- Tranebaerg L. Genetics of congenital hearing impairment: a clinical approach. Int. J. Audiol. 2008;47:535–545. - PubMed
-
- Morton C.C., Nance W.E. Newborn hearing screening—a silent revolution. N. Engl. J. Med. 2006;354:2151–2164. - PubMed
-
- Collin R.W., Littink K.W., Klevering B.J., van den Born L.I., Koenekoop R.K., Zonneveld M.N., Blokland E.A., Strom T.M., Hoyng C.B., den Hollander A.I., Cremers F.P. Identification of a 2 Mb human ortholog of Drosophila eyes shut/spacemaker that is mutated in patients with retinitis pigmentosa. Am. J. Hum. Genet. 2008;83:594–603. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

