Two intramolecular isopeptide bonds are identified in the crystal structure of the Streptococcus gordonii SspB C-terminal domain
- PMID: 20138058
- PMCID: PMC2849992
- DOI: 10.1016/j.jmb.2010.01.065
Two intramolecular isopeptide bonds are identified in the crystal structure of the Streptococcus gordonii SspB C-terminal domain
Abstract
Streptococcus gordonii is a primary colonizer and is involved in the formation of dental plaque. This bacterium expresses several surface proteins. One of them is the adhesin SspB, which is a member of the Antigen I/II family of proteins. SspB is a large multi-domain protein that has interactions with surface molecules on other bacteria and on host cells, and is thus a key factor in the formation of biofilms. Here, we report the crystal structure of a truncated form of the SspB C-terminal domain, solved by single-wavelength anomalous dispersion to 1.5 A resolution. The structure represents the first of a C-terminal domain from a streptococcal Antigen I/II protein and is comprised of two structurally related beta-sandwich domains, C2 and C3, both with a Ca(2+) bound in equivalent positions. In each of the domains, a covalent isopeptide bond is observed between a lysine and an asparagine, a feature that is believed to be a common stabilization mechanism in Gram-positive surface proteins. S. gordonii biofilms contain attachment sites for the periodontal pathogen Porphyromonas gingivalis and the SspB C-terminal domain has been shown to have one such recognition motif, the SspB adherence region. The motif protrudes from the protein, and serves as a handle for attachment. The structure suggests several additional putative binding surfaces, and other binding clefts may be created when the full-length protein is folded.
Copyright 2010 Elsevier Ltd. All rights reserved.
Figures









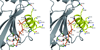
Similar articles
-
Crystal structure of the variable domain of the Streptococcus gordonii surface protein SspB.Protein Sci. 2009 Sep;18(9):1896-905. doi: 10.1002/pro.200. Protein Sci. 2009. PMID: 19609934 Free PMC article.
-
Structural and functional analysis of the C-terminal region of Streptococcus gordonii SspB.Acta Crystallogr D Struct Biol. 2021 Sep 1;77(Pt 9):1206-1215. doi: 10.1107/S2059798321008135. Epub 2021 Aug 27. Acta Crystallogr D Struct Biol. 2021. PMID: 34473090 Free PMC article.
-
A crystallizable form of the Streptococcus gordonii surface antigen SspB C-domain obtained by limited proteolysis.Acta Crystallogr Sect F Struct Biol Cryst Commun. 2009 Jul 1;65(Pt 7):712-4. doi: 10.1107/S1744309109021046. Epub 2009 Jun 27. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2009. PMID: 19574647 Free PMC article.
-
The changing faces of Streptococcus antigen I/II polypeptide family adhesins.Mol Microbiol. 2010 Jul;77(2):276-86. doi: 10.1111/j.1365-2958.2010.07212.x. Epub 2010 May 24. Mol Microbiol. 2010. PMID: 20497507 Free PMC article. Review.
-
[Adhesins of oral streptococci].Nihon Saikingaku Zasshi. 2013;68(2):283-93. doi: 10.3412/jsb.68.283. Nihon Saikingaku Zasshi. 2013. PMID: 23727707 Review. Japanese.
Cited by
-
Microbial interactions in building of communities.Mol Oral Microbiol. 2013 Apr;28(2):83-101. doi: 10.1111/omi.12012. Epub 2012 Dec 17. Mol Oral Microbiol. 2013. PMID: 23253299 Free PMC article. Review.
-
A slow-forming isopeptide bond in the structure of the major pilin SpaD from Corynebacterium diphtheriae has implications for pilus assembly.Acta Crystallogr D Biol Crystallogr. 2014 May;70(Pt 5):1190-201. doi: 10.1107/S1399004714001400. Epub 2014 Apr 26. Acta Crystallogr D Biol Crystallogr. 2014. PMID: 24816089 Free PMC article.
-
Community signalling between Streptococcus gordonii and Porphyromonas gingivalis is controlled by the transcriptional regulator CdhR.Mol Microbiol. 2010 Dec;78(6):1510-22. doi: 10.1111/j.1365-2958.2010.07420.x. Epub 2010 Oct 26. Mol Microbiol. 2010. PMID: 21143321 Free PMC article.
-
Structural dissection and in vivo effectiveness of a peptide inhibitor of Porphyromonas gingivalis adherence to Streptococcus gordonii.Infect Immun. 2011 Jan;79(1):67-74. doi: 10.1128/IAI.00361-10. Epub 2010 Nov 1. Infect Immun. 2011. PMID: 21041492 Free PMC article.
-
Stick to your gums: mechanisms of oral microbial adherence.J Dent Res. 2011 Nov;90(11):1271-8. doi: 10.1177/0022034511399096. Epub 2011 Feb 18. J Dent Res. 2011. PMID: 21335541 Free PMC article. Review.
References
-
- Jenkinson HF, Lamont RJ. Oral microbial communities in sickness and in health. Trends Microbiol. 2005;13:589–595. - PubMed
-
- Nyvad B, Kilian M. Comparison of the initial streptococcal microflora on dental enamel in caries-active and in caries-inactive individuals. Caries Res. 1990;24:267–272. - PubMed
-
- Jenkinson HF, Lamont RJ. Streptococcal adhesion and colonization. Crit. Rev. Oral Biol. Med. 1997;8:175–200. - PubMed
-
- Rosan B, Lamont RJ. Dental plaque formation. Microbes Infect. 2000;2:1599–1607. - PubMed
-
- Douglas CW, Heath J, Hampton KK, Preston FE. Identity of viridans streptococci isolated from cases of infective endocarditis. J. Med. Microbiol. 1993;39:179–182. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

