Critical requirement of GABPalpha for normal T cell development
- PMID: 20139079
- PMCID: PMC2856223
- DOI: 10.1074/jbc.M109.088740
Critical requirement of GABPalpha for normal T cell development
Abstract
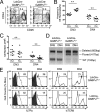
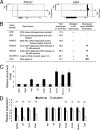
GA binding protein (GABP) consists of GABPalpha and GABPbeta subunits. GABPalpha is a member of Ets family transcription factors and binds DNA via its conserved Ets domain, whereas GABPbeta does not bind DNA but possesses transactivation activity. In T cells, GABP has been demonstrated to regulate the gene expression of interleukin-7 receptor alpha chain (IL-7Ralpha) and postulated to be critical in T cell development. To directly investigate its function in early thymocyte development, we used GABPalpha conditional knock-out mice where the exons encoding the Ets DNA-binding domain are flanked with LoxP sites. Ablation of GABPalpha with the Lck-Cre transgene greatly diminished thymic cellularity, blocked thymocyte development at the double negative 3 (DN3) stage, and resulted in reduced expression of T cell receptor (TCR) beta chain in DN4 thymocytes. By chromatin immunoprecipitation, we demonstrated in DN thymocytes that GABPalpha is associated with transcription initiation sites of genes encoding key molecules in TCR rearrangements. Among these GABP-associated genes, knockdown of GABPalpha expression by RNA interference diminished expression of DNA ligase IV, Artemis, and Ku80 components in DNA-dependent protein kinase complex. Interestingly, forced expression of prearranged TCR but not IL-7Ralpha can alleviate the DN3 block in GABPalpha-targeted mice. Our observations collectively indicate that in addition to regulating IL-7Ralpha expression, GABP is critically required for TCR rearrangements and hence normal T cell development.
Figures




Similar articles
-
GABP controls a critical transcription regulatory module that is essential for maintenance and differentiation of hematopoietic stem/progenitor cells.Blood. 2011 Feb 17;117(7):2166-78. doi: 10.1182/blood-2010-09-306563. Epub 2010 Dec 7. Blood. 2011. PMID: 21139080 Free PMC article.
-
The transcription factor GABP is a critical regulator of B lymphocyte development.Immunity. 2007 Apr;26(4):421-31. doi: 10.1016/j.immuni.2007.03.010. Immunity. 2007. PMID: 17442597
-
The ETS transcription factor GABPalpha is essential for early embryogenesis.Mol Cell Biol. 2004 Jul;24(13):5844-9. doi: 10.1128/MCB.24.13.5844-5849.2004. Mol Cell Biol. 2004. PMID: 15199140 Free PMC article.
-
At the crossroads: diverse roles of early thymocyte transcriptional regulators.Immunol Rev. 2006 Feb;209:191-211. doi: 10.1111/j.0105-2896.2006.00352.x. Immunol Rev. 2006. PMID: 16448544 Review.
-
GA-binding protein transcription factor: a review of GABP as an integrator of intracellular signaling and protein-protein interactions.Blood Cells Mol Dis. 2004 Jan-Feb;32(1):143-54. doi: 10.1016/j.bcmd.2003.09.005. Blood Cells Mol Dis. 2004. PMID: 14757430 Review.
Cited by
-
Genome-wide analysis reveals conserved and divergent features of Notch1/RBPJ binding in human and murine T-lymphoblastic leukemia cells.Proc Natl Acad Sci U S A. 2011 Sep 6;108(36):14908-13. doi: 10.1073/pnas.1109023108. Epub 2011 Jul 7. Proc Natl Acad Sci U S A. 2011. PMID: 21737748 Free PMC article.
-
Sense-antisense gene-pairs in breast cancer and associated pathological pathways.Oncotarget. 2015 Dec 8;6(39):42197-221. doi: 10.18632/oncotarget.6255. Oncotarget. 2015. PMID: 26517092 Free PMC article.
-
The possible functions of duplicated ets (GGAA) motifs located near transcription start sites of various human genes.Cell Mol Life Sci. 2011 Jun;68(12):2039-51. doi: 10.1007/s00018-011-0674-x. Epub 2011 Apr 3. Cell Mol Life Sci. 2011. PMID: 21461879 Free PMC article.
-
GABP controls a critical transcription regulatory module that is essential for maintenance and differentiation of hematopoietic stem/progenitor cells.Blood. 2011 Feb 17;117(7):2166-78. doi: 10.1182/blood-2010-09-306563. Epub 2010 Dec 7. Blood. 2011. PMID: 21139080 Free PMC article.
-
Differentiation and persistence of memory CD8(+) T cells depend on T cell factor 1.Immunity. 2010 Aug 27;33(2):229-40. doi: 10.1016/j.immuni.2010.08.002. Immunity. 2010. PMID: 20727791 Free PMC article.
References
-
- Bhandoola A., von Boehmer H., Petrie H. T., Zúñiga-Pflücker J. C. (2007) Immunity 26, 678–689 - PubMed
-
- Rothenberg E. V., Taghon T. (2005) Annu. Rev. Immunol. 23, 601–649 - PubMed
-
- Scott E. W., Simon M. C., Anastasi J., Singh H. (1994) Science 265, 1573–1577 - PubMed
-
- Georgopoulos K. (2002) Nat. Rev. Immunol. 2, 162–174 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

