Molecular basis of the waxy endosperm starch phenotype in broomcorn millet (Panicum miliaceum L.)
- PMID: 20139147
- PMCID: PMC2884200
- DOI: 10.1093/molbev/msq040
Molecular basis of the waxy endosperm starch phenotype in broomcorn millet (Panicum miliaceum L.)
Abstract
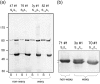
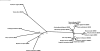
Waxy varieties of the tetraploid cereal broomcorn millet (Panicum miliaceum L.) have endosperm starch granules lacking detectable amylose. This study investigated the basis of this phenotype using molecular and biochemical methods. Iodine staining of starch granules in 72 plants from 38 landrace accessions found 58 nonwaxy and 14 waxy phenotype plants. All waxy types were in plants from Chinese and Korean accessions, a distribution similar to that of the waxy phenotype in other cereals. Granule-bound starch synthase I (GBSSI) protein was present in the endosperm of both nonwaxy and waxy individuals, but waxy types had little or no granule-bound starch synthase activity compared with the wild types. Sequencing of the GBSSI (Waxy) gene showed that this gene is present in two different forms (L and S) in P. miliaceum, which probably represent homeologues derived from two distinct diploid ancestors. Protein products of both these forms are present in starch granules. We identified three polymorphisms in the exon sequence coding for mature GBSSI peptides. A 15-bp deletion has occurred in the S type GBSSI, resulting in the loss of five amino acids from glucosyl transferase domain 1 (GTD1). The second GBSSI type (L) shows two sequence polymorphisms. One is the insertion of an adenine residue that causes a reading frameshift, and the second causes a cysteine-tyrosine amino acid polymorphism. These mutations appear to have occurred in parallel from the ancestral allele, resulting in three GBSSI-L alleles in total. Five of the six possible genotype combinations of the S and L alleles were observed. The deletion in the GBSSI-S gene causes loss of protein activity, and there was 100% correspondence between this deletion and the waxy phenotype. The frameshift mutation in the L gene results in the loss of L-type protein from starch granules. The L isoform with the tyrosine residue is present in starch granules but is nonfunctional. This loss of function may result from the substitution of tyrosine for cysteine, although it could not be determined whether the cysteine isoform of L represents the functional type. This is the first characterization of mutations that occur in combination in a functionally polyploid species to give a fully waxy phenotype.
Figures
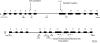

References
-
- Abascal F, Zardoya R, Posada D. ProtTest: selection of bestfit models of protein evolution. Bioinformatics. 2005;21:2104–2105. - PubMed
-
- Baltensperger DD. Foxtail and Proso millet. In: Janick J, editor. Progress in new crops. ASHS Press: Alexandria (VA); 1996. pp. 182–190.
-
- Bouchenak-Khelladi Y, Salamin N, Savolainen V, Forest F, van der Bank M, Chase MW, Hodkinson TR. Large multi-gene phylogenetic trees of the grasses (Poaceae): progress towards complete tribal and generic level sampling. Mol Phylogenet Evol. 2008;47:488–505. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

