The DMM complex prevents spreading of DNA methylation from transposons to nearby genes in Neurospora crassa
- PMID: 20139222
- PMCID: PMC2827840
- DOI: 10.1101/gad.1893210
The DMM complex prevents spreading of DNA methylation from transposons to nearby genes in Neurospora crassa
Abstract
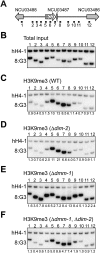
Transposable elements are common in genomes and must be controlled. Many organisms use DNA methylation to silence such selfish DNA, but the mechanisms that restrict the methylation to appropriate regions are largely unknown. We identified a JmjC domain protein in Neurospora, DNA METHYLATION MODULATOR-1 (DMM-1), that prevents aberrant spreading of DNA and histone H3K9 methylation from inactivated transposons into nearby genes. Mutation of a conserved residue within the JmjC Fe(II)-binding site abolished dmm-1 function, as did mutations in conserved cysteine-rich domains. Mutants defective only in dmm-1 mutants grow poorly, but growth is restored by reduction or elimination of DNA methylation using the drug 5-azacytosine or by mutation of the DNA methyltransferase gene dim-2. DMM-1 relies on an associated protein, DMM-2, which bears a DNA-binding motif, for localization and proper function. HP1 is required to recruit the DMM complex to the edges of methylated regions.
Figures

Similar articles
-
The methylated component of the Neurospora crassa genome.Nature. 2003 Apr 24;422(6934):893-7. doi: 10.1038/nature01564. Nature. 2003. PMID: 12712205
-
DNA methylation and normal chromosome behavior in Neurospora depend on five components of a histone methyltransferase complex, DCDC.PLoS Genet. 2010 Nov 4;6(11):e1001196. doi: 10.1371/journal.pgen.1001196. PLoS Genet. 2010. PMID: 21079689 Free PMC article.
-
Direct interaction between DNA methyltransferase DIM-2 and HP1 is required for DNA methylation in Neurospora crassa.Mol Cell Biol. 2008 Oct;28(19):6044-55. doi: 10.1128/MCB.00823-08. Epub 2008 Aug 4. Mol Cell Biol. 2008. PMID: 18678653 Free PMC article.
-
DNA methylation and the formation of heterochromatin in Neurospora crassa.Heredity (Edinb). 2010 Jul;105(1):38-44. doi: 10.1038/hdy.2010.44. Epub 2010 Apr 21. Heredity (Edinb). 2010. PMID: 20407471 Review.
-
Genetic and epigenetic inactivation of repetitive sequences in Neurospora crassa: RIP, DNA methylation, and quelling.Curr Top Microbiol Immunol. 1995;197:165-77. doi: 10.1007/978-3-642-79145-1_11. Curr Top Microbiol Immunol. 1995. PMID: 7493491 Review. No abstract available.
Cited by
-
An H3K9 methylation-dependent protein interaction regulates the non-enzymatic functions of a putative histone demethylase.Elife. 2020 Mar 20;9:e53155. doi: 10.7554/eLife.53155. Elife. 2020. PMID: 32195666 Free PMC article.
-
Induction of H3K9me3 and DNA methylation by tethered heterochromatin factors in Neurospora crassa.Proc Natl Acad Sci U S A. 2017 Nov 7;114(45):E9598-E9607. doi: 10.1073/pnas.1715049114. Epub 2017 Oct 23. Proc Natl Acad Sci U S A. 2017. PMID: 29078403 Free PMC article.
-
Coevolution of the CDCA7-HELLS ICF-related nucleosome remodeling complex and DNA methyltransferases.Elife. 2023 Sep 28;12:RP86721. doi: 10.7554/eLife.86721. Elife. 2023. PMID: 37769127 Free PMC article.
-
Conserved gene regulatory function of the carboxy-terminal domain of dictyostelid C-module-binding factor.Eukaryot Cell. 2013 Mar;12(3):460-8. doi: 10.1128/EC.00329-12. Epub 2013 Jan 25. Eukaryot Cell. 2013. PMID: 23355006 Free PMC article.
-
KdmA, a histone H3 demethylase with bipartite function, differentially regulates primary and secondary metabolism in Aspergillus nidulans.Mol Microbiol. 2015 May;96(4):839-60. doi: 10.1111/mmi.12977. Epub 2015 Apr 11. Mol Microbiol. 2015. PMID: 25712266 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
