Complete HOX cluster characterization of the coelacanth provides further evidence for slow evolution of its genome
- PMID: 20139301
- PMCID: PMC2840454
- DOI: 10.1073/pnas.0914312107
Complete HOX cluster characterization of the coelacanth provides further evidence for slow evolution of its genome
Abstract
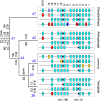
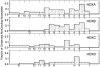
The living coelacanth is a lobe-finned fish that represents an early evolutionary departure from the lineage that led to land vertebrates, and is of extreme interest scientifically. It has changed very little in appearance from fossilized coelacanths of the Cretaceous (150 to 65 million years ago), and is often referred to as a "living fossil." An important general question is whether long-term stasis in morphological evolution is associated with stasis in genome evolution. To this end we have used targeted genome sequencing for acquiring 1,612,752 bp of high quality finished sequence encompassing the four HOX clusters of the Indonesian coelacanth Latimeria menadoensis. Detailed analyses were carried out on genomic structure, gene and repeat contents, conserved noncoding regions, and relative rates of sequence evolution in both coding and noncoding tracts. Our results demonstrate conclusively that the coelacanth HOX clusters are evolving comparatively slowly and that this taxon should serve as a viable outgroup for interpretation of the genomes of tetrapod species.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Balon EK, Bruton MN, Fricke H. A fiftieth anniversary reflection on the living coelacanth, Latimeria chalumnae: some new interpretations of its natural history and conservation status. Environ Biol Fishes. 1988;23:241–280.
-
- Danke J, et al. Genome resource for the indonesian coelacanth, Latimeria menadoensis . J Exp Zoolog A Comp Exp Biol. 2004;301:228–234. - PubMed
-
- Shashikant C, Bolanowski SA, Danke J, Amemiya CT. Hoxc8 early enhancer of the indonesian coelacanth, Latimeria menadoensis . J Exp Zoolog B Mol Dev Evol. 2004;302:557–563. - PubMed
-
- Carroll RL. Vertebrate paleontology and evolution. New York: H. Freeman and Co.; 1988.
-
- Takezaki N, Figueroa F, Zaleska-Rutczynska Z, Takahata N, Klein J. The phylogenetic relationship of tetrapod, coelacanth, and lungfish revealed by the sequences of forty-four nuclear genes. Mol Biol Evol. 2004;21:1512–1524. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources

