Genomic code for Sox10 activation reveals a key regulatory enhancer for cranial neural crest
- PMID: 20139305
- PMCID: PMC2840498
- DOI: 10.1073/pnas.0906596107
Genomic code for Sox10 activation reveals a key regulatory enhancer for cranial neural crest
Abstract
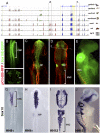
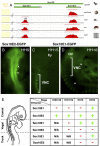
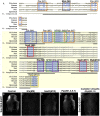
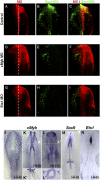
The neural crest is a multipotent, stem cell-like population that migrates extensively in the embryo and forms a wide array of derivatives, ranging from neurons to melanocytes and cartilage. Analyses of the gene regulatory network driving neural crest development revealed Sox10 as one of the earliest neural crest-specifying genes, cell-autonomously driving delamination and directly regulating numerous downstream effectors and differentiation gene batteries. In search of direct inputs to the neural crest specifier module, we dissected the chick Sox10 genomic region and isolated two downstream regulatory regions with distinct spatiotemporal activity. A unique element, Sox10E2 represents the earliest-acting neural crest cis-regulatory element, critical for initiating Sox10 expression in newly formed cranial, but not vagal and trunk neural crest. A second element, Sox10E1, acts in later migrating vagal and trunk crest cells. Deep characterization of Sox10E2 reveals Sox9, Ets1, and cMyb as direct inputs mediating enhancer activity. ChIP, DNA-pull down, and gel-shift assays demonstrate their direct binding to the Sox10E2 enhancer in vivo, whereas mutation of their corresponding binding sites, or inactivation of the three upstream regulators, abolishes both reporter and endogenous Sox10 expression. Using cis-regulatory analysis as a tool, our study makes critical connections within the neural crest gene regulatory network, thus being unique in establishing a direct link of upstream effectors to a key neural crest specifier.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
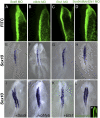
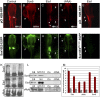
References
-
- Martinsen BJ, Bronner-Fraser M. Neural crest specification regulated by the helix-loop-helix repressor Id2. Science. 1998;281:988–991. - PubMed
-
- Tahtakran SA, Selleck MA. Ets-1 expression is associated with cranial neural crest migration and vasculogenesis in the chick embryo. Gene Expr Patterns. 2003;3:455–458. - PubMed
-
- Meulemans D, Bronner-Fraser M. Gene-regulatory interactions in neural crest evolution and development. Dev Cell. 2004;7:291–299. - PubMed
-
- Sauka-Spengler T, Bronner-Fraser M. Development and evolution of the migratory neural crest: a gene regulatory perspective. Curr Opin Genet Dev. 2006;16:360–366. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

