A real-time assay for CpG-specific cytosine-C5 methyltransferase activity
- PMID: 20139415
- PMCID: PMC2875032
- DOI: 10.1093/nar/gkq047
A real-time assay for CpG-specific cytosine-C5 methyltransferase activity
Abstract
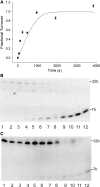
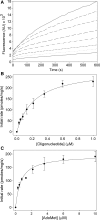
A real-time assay for CpG-specific cytosine-C5 methyltransferase activity has been developed. The assay applies a break light oligonucleotide in which the methylation of an unmethylated 5'-CG-3' site is enzymatically coupled to the development of a fluorescent signal. This sensitive assay can measure rates of DNA methylation down to 0.34 +/- 0.06 fmol/s. The assay is reproducible, with a coefficient of variation over six independent measurements of 4.5%. Product concentration was accurately measured from fluorescence signals using a linear calibration curve, which achieved a goodness of fit (R(2)) above 0.98. The oligonucleotide substrate contains three C5-methylated cytosine residues and one unmethylated 5'-CG-3' site. Methylation yields an oligonucleotide containing the optimal substrate for the restriction enzyme GlaI. Cleavage of the fully methylated oligonucleotide leads to separation of fluorophore from quencher, giving a proportional increase in fluorescence. This method has been used to assay activity of DNMT1, the principle maintenance methyltransferase in human cells, and for the kinetic characterization of the bacterial cytosine-C5 methyltransferase M.SssI. The assay has been shown to be suitable for the real-time monitoring of DNMT1 activity in a high-throughput format, with low background signal and the ability to obtain linear rates of methylation over long periods, making this a promising method of high-throughput screening for inhibitors.
Figures

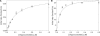



Similar articles
-
Kinetic analysis of Yersinia pestis DNA adenine methyltransferase activity using a hemimethylated molecular break light oligonucleotide.PLoS One. 2007 Aug 29;2(8):e801. doi: 10.1371/journal.pone.0000801. PLoS One. 2007. PMID: 17726531 Free PMC article.
-
Fluorescence-based high-throughput assay for human DNA (cytosine-5)-methyltransferase 1.Anal Biochem. 2010 Jun 1;401(1):168-72. doi: 10.1016/j.ab.2010.02.032. Epub 2010 Mar 1. Anal Biochem. 2010. PMID: 20197058 Free PMC article.
-
The Dnmt1 DNA-(cytosine-C5)-methyltransferase methylates DNA processively with high preference for hemimethylated target sites.J Biol Chem. 2004 Nov 12;279(46):48350-9. doi: 10.1074/jbc.M403427200. Epub 2004 Aug 31. J Biol Chem. 2004. PMID: 15339928
-
Mammalian cytosine DNA methyltransferase Dnmt1: enzymatic mechanism, novel mechanism-based inhibitors, and RNA-directed DNA methylation.Curr Med Chem. 2008;15(1):92-106. doi: 10.2174/092986708783330700. Curr Med Chem. 2008. PMID: 18220765 Review.
-
DNA Methyltransferases in Mammalian Oocytes.Results Probl Cell Differ. 2017;63:211-222. doi: 10.1007/978-3-319-60855-6_10. Results Probl Cell Differ. 2017. PMID: 28779320 Review.
Cited by
-
Z-DNA as a Tool for Nuclease-Free DNA Methyltransferase Assay.Int J Mol Sci. 2021 Nov 5;22(21):11990. doi: 10.3390/ijms222111990. Int J Mol Sci. 2021. PMID: 34769422 Free PMC article.
-
An integrated-molecular-beacon based multiple exponential strand displacement amplification strategy for ultrasensitive detection of DNA methyltransferase activity.Chem Sci. 2018 Dec 20;10(8):2290-2297. doi: 10.1039/c8sc05102j. eCollection 2019 Feb 28. Chem Sci. 2018. PMID: 30881654 Free PMC article.
-
Expression and purification of the modification-dependent restriction enzyme BisI and its homologous enzymes.Sci Rep. 2016 Jun 29;6:28579. doi: 10.1038/srep28579. Sci Rep. 2016. PMID: 27353146 Free PMC article.
-
HpaII-assisted and linear amplification-enhanced isothermal exponential amplification fluorescent strategy for rapid and sensitive detection of DNA methyltransferase activity.Anal Bioanal Chem. 2023 May;415(12):2271-2280. doi: 10.1007/s00216-023-04647-1. Epub 2023 Mar 24. Anal Bioanal Chem. 2023. PMID: 36961574
-
Electrochemical assay for the signal-on detection of human DNA methyltransferase activity.J Am Chem Soc. 2013 Nov 6;135(44):16632-40. doi: 10.1021/ja4085918. J Am Chem Soc. 2013. PMID: 24164112 Free PMC article.
References
-
- Colot V, Rossignol JL. Eukaryotic DNA methylation as an evolutionary device. Bioessays. 1999;21:402–411. - PubMed
-
- Johnson TB, Coghill RD. Researches on pyrimidines C111. The discovery of 5-methylcytosine in tuberculinic acid, the nucleic acid of the tubercle bacillus. J. Am. Chem. Soc. 1925;47:2838–2844.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

