Trans-synaptic adhesions between netrin-G ligand-3 (NGL-3) and receptor tyrosine phosphatases LAR, protein-tyrosine phosphatase delta (PTPdelta), and PTPsigma via specific domains regulate excitatory synapse formation
- PMID: 20139422
- PMCID: PMC2859559
- DOI: 10.1074/jbc.M109.061127
Trans-synaptic adhesions between netrin-G ligand-3 (NGL-3) and receptor tyrosine phosphatases LAR, protein-tyrosine phosphatase delta (PTPdelta), and PTPsigma via specific domains regulate excitatory synapse formation
Abstract
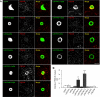
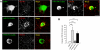
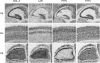
Synaptic cell adhesion molecules regulate various steps of synapse formation. The trans-synaptic adhesion between postsynaptic NGL-3 (for netrin-G ligand-3) and presynaptic LAR (for leukocyte antigen-related) regulates excitatory synapse formation in a bidirectional manner. However, little is known about the molecular details of the NGL-3-LAR adhesion and whether two additional LAR family proteins, protein-tyrosine phosphatase delta (PTPdelta), and PTPsigma, also interact with NGL-3 and are involved in synapse formation. We report here that the leucine-rich repeat (LRR) domain of NGL-3, containing nine LRRs, interacts with the first two fibronectin III (FNIII) domains of LAR to induce bidirectional synapse formation. Moreover, Gln-96 in the first LRR motif of NGL-3 is critical for LAR binding and induction of presynaptic differentiation. PTPdelta and PTPsigma also interact with NGL-3 via their first two FNIII domains. These two interactions promote synapse formation in a different manner; the PTPsigma-NGL-3 interaction promotes synapse formation in a bidirectional manner, whereas the PTPdelta-NGL-3 interaction instructs only presynaptic differentiation in a unidirectional manner. mRNAs encoding LAR family proteins display overlapping and differential expression patterns in various brain regions. These results suggest that trans-synaptic adhesion between NGL-3 and the three LAR family proteins regulates excitatory synapse formation in shared and distinct neural circuits.
Figures

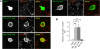
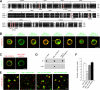
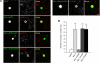
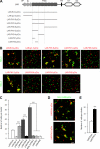


Similar articles
-
Trans-synaptic adhesion between NGL-3 and LAR regulates the formation of excitatory synapses.Nat Neurosci. 2009 Apr;12(4):428-37. doi: 10.1038/nn.2279. Epub 2009 Mar 1. Nat Neurosci. 2009. PMID: 19252495
-
Trans-induced cis interaction in the tripartite NGL-1, netrin-G1 and LAR adhesion complex promotes development of excitatory synapses.J Cell Sci. 2013 Nov 1;126(Pt 21):4926-38. doi: 10.1242/jcs.129718. Epub 2013 Aug 28. J Cell Sci. 2013. PMID: 23986473
-
LAR-RPTPs Directly Interact with Neurexins to Coordinate Bidirectional Assembly of Molecular Machineries.J Neurosci. 2020 Oct 28;40(44):8438-8462. doi: 10.1523/JNEUROSCI.1091-20.2020. Epub 2020 Oct 9. J Neurosci. 2020. PMID: 33037075 Free PMC article.
-
LAR-RPTPs: synaptic adhesion molecules that shape synapse development.Trends Cell Biol. 2013 Oct;23(10):465-75. doi: 10.1016/j.tcb.2013.07.004. Epub 2013 Aug 3. Trends Cell Biol. 2013. PMID: 23916315 Review.
-
The NGL family of leucine-rich repeat-containing synaptic adhesion molecules.Mol Cell Neurosci. 2009 Sep;42(1):1-10. doi: 10.1016/j.mcn.2009.05.008. Epub 2009 May 23. Mol Cell Neurosci. 2009. PMID: 19467332 Review.
Cited by
-
Mechanisms of splicing-dependent trans-synaptic adhesion by PTPδ-IL1RAPL1/IL-1RAcP for synaptic differentiation.Nat Commun. 2015 Apr 24;6:6926. doi: 10.1038/ncomms7926. Nat Commun. 2015. PMID: 25908590 Free PMC article.
-
Receptor protein tyrosine phosphatases and cancer: new insights from structural biology.Cell Adh Migr. 2012 Jul-Aug;6(4):356-64. doi: 10.4161/cam.21242. Epub 2012 Jul 1. Cell Adh Migr. 2012. PMID: 22796942 Free PMC article. Review.
-
Receptor-type tyrosine phosphatase ligands: looking for the needle in the haystack.FEBS J. 2013 Jan;280(2):388-400. doi: 10.1111/j.1742-4658.2012.08653.x. Epub 2012 Jul 5. FEBS J. 2013. PMID: 22682003 Free PMC article. Review.
-
Structure-activity studies of PTPRD phosphatase inhibitors identify a 7-cyclopentymethoxy illudalic acid analog candidate for development.Biochem Pharmacol. 2022 Jan;195:114868. doi: 10.1016/j.bcp.2021.114868. Epub 2021 Dec 2. Biochem Pharmacol. 2022. PMID: 34863978 Free PMC article.
-
Postsynaptic TrkC and presynaptic PTPσ function as a bidirectional excitatory synaptic organizing complex.Neuron. 2011 Jan 27;69(2):287-303. doi: 10.1016/j.neuron.2010.12.024. Neuron. 2011. PMID: 21262467 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

