Snf1 dependence of peroxisomal gene expression is mediated by Adr1
- PMID: 20139423
- PMCID: PMC2856278
- DOI: 10.1074/jbc.M109.079848
Snf1 dependence of peroxisomal gene expression is mediated by Adr1
Abstract
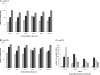
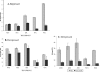
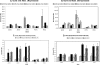
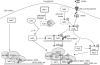
Eukaryotes utilize fatty acids by beta-oxidation, which occurs in the mitochondria and peroxisomes in higher organisms and in the peroxisomes in yeast. The AMP-activated protein kinase regulates this process in mammalian cells, and its homolog Snf1, together with the transcription factors Adr1, Oaf1, and Pip2, regulates peroxisome proliferation and beta-oxidation in yeast. A constitutive allele of Adr1 (Adr1(c)) lacking the glucose- and Snf1-regulated phosphorylation substrate Ser-230 was found to be Snf1-independent for regulation of peroxisomal genes. In addition, it could compensate for and even suppress the requirement for Oaf1 or Pip2 for gene induction. Peroxisomal genes were found to be regulated by oleate in the presence of glucose, as long as Adr1(c) was expressed, suggesting that the Oaf1/Pip2 heterodimer is Snf1-independent. Consistent with this observation, Oaf1 binding to promoters in the presence of oleate was not reduced in a snf1Delta strain. Exploring the mechanism by which Adr1(c) permits Snf1-independent peroxisomal gene induction, we found that strength of promoter binding did not correlate with transcription, suggesting that stable binding is not a prerequisite for enhanced transcription. Instead, enhanced transcriptional activation and suppression of Oaf1, Pip2, and Snf1 by Adr1(c) may be related to the ability of Adr1(c) to suppress the requirement for and enhance the recruitment of transcriptional coactivators in a promoter- and growth medium-dependent manner.
Figures


Similar articles
-
14-3-3 (Bmh) proteins regulate combinatorial transcription following RNA polymerase II recruitment by binding at Adr1-dependent promoters in Saccharomyces cerevisiae.Mol Cell Biol. 2013 Feb;33(4):712-24. doi: 10.1128/MCB.01226-12. Epub 2012 Dec 3. Mol Cell Biol. 2013. PMID: 23207903 Free PMC article.
-
Snf1 controls the activity of adr1 through dephosphorylation of Ser230.Genetics. 2009 Jul;182(3):735-45. doi: 10.1534/genetics.109.103432. Epub 2009 Apr 27. Genetics. 2009. PMID: 19398770 Free PMC article.
-
Yeast 14-3-3 protein functions as a comodulator of transcription by inhibiting coactivator functions.J Biol Chem. 2014 Dec 19;289(51):35542-60. doi: 10.1074/jbc.M114.592287. Epub 2014 Oct 29. J Biol Chem. 2014. PMID: 25355315 Free PMC article.
-
Transcriptional control of nonfermentative metabolism in the yeast Saccharomyces cerevisiae.Curr Genet. 2003 Jun;43(3):139-60. doi: 10.1007/s00294-003-0381-8. Epub 2003 Apr 25. Curr Genet. 2003. PMID: 12715202 Review.
-
The biochemistry of oleate induction: transcriptional upregulation and peroxisome proliferation.Biochim Biophys Acta. 2006 Dec;1763(12):1392-402. doi: 10.1016/j.bbamcr.2006.07.011. Epub 2006 Jul 26. Biochim Biophys Acta. 2006. PMID: 16949166 Review.
Cited by
-
PhosphoChain: a novel algorithm to predict kinase and phosphatase networks from high-throughput expression data.Bioinformatics. 2013 Oct 1;29(19):2435-44. doi: 10.1093/bioinformatics/btt387. Epub 2013 Jul 5. Bioinformatics. 2013. PMID: 23832245 Free PMC article.
-
Mitochondria-cytosol-nucleus crosstalk: learning from Saccharomyces cerevisiae.FEMS Yeast Res. 2018 Dec 1;18(8):foy088. doi: 10.1093/femsyr/foy088. FEMS Yeast Res. 2018. PMID: 30165482 Free PMC article. Review.
-
Mechanism of enhanced salt tolerance in Saccharomyces cerevisiae by CRZ1 overexpression.Sci Rep. 2024 Oct 2;14(1):22875. doi: 10.1038/s41598-024-74174-1. Sci Rep. 2024. PMID: 39358483 Free PMC article.
-
Activator-independent transcription of Snf1-dependent genes in mutants lacking histone tails.Mol Microbiol. 2011 Apr;80(2):407-22. doi: 10.1111/j.1365-2958.2011.07583.x. Epub 2011 Mar 1. Mol Microbiol. 2011. PMID: 21338416 Free PMC article.
-
14-3-3 (Bmh) proteins regulate combinatorial transcription following RNA polymerase II recruitment by binding at Adr1-dependent promoters in Saccharomyces cerevisiae.Mol Cell Biol. 2013 Feb;33(4):712-24. doi: 10.1128/MCB.01226-12. Epub 2012 Dec 3. Mol Cell Biol. 2013. PMID: 23207903 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

