Chemical phylogenetics of histone deacetylases
- PMID: 20139990
- PMCID: PMC2822059
- DOI: 10.1038/nchembio.313
Chemical phylogenetics of histone deacetylases
Abstract
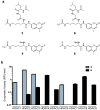
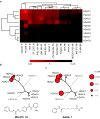
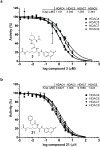
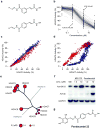
The broad study of histone deacetylases in chemistry, biology and medicine relies on tool compounds to derive mechanistic insights. A phylogenetic analysis of class I and II histone deacetylases (HDACs) as targets of a comprehensive, structurally diverse panel of inhibitors revealed unexpected isoform selectivity even among compounds widely perceived as nonselective. The synthesis and study of a focused library of cinnamic hydroxamates allowed the identification of, to our knowledge, the first nonselective HDAC inhibitor. These data will guide a more informed use of HDAC inhibitors as chemical probes and therapeutic agents.
Figures
References
-
- Minucci S, Pelicci PG. Histone deacetylase inhibitors and the promise of epigenetic (and more) treatments for cancer. Nat Rev Cancer. 2006;6:38–51. - PubMed
-
- Lee KK, Workman JL. Histone acetyltransferase complexes: one size doesn’t fit all. Nat Rev Mol Cell Biol. 2007;8:284–295. - PubMed
-
- Choudhary C, et al. Lysine Acetylation Targets Protein Complexes and Co-Regulates Major Cellular Functions. Science. 2009;325:834–840. - PubMed
-
- Gregoretti IV, Lee YM, Goodson HV. Molecular evolution of the histone deacetylase family: functional implications of phylogenetic analysis. J Mol Biol. 2004;338:17–31. - PubMed
Associated data
- PubChem-Substance/87226482
- PubChem-Substance/87226502
- PubChem-Substance/87226493
- PubChem-Substance/87226494
- PubChem-Substance/87226483
- PubChem-Substance/87226495
- PubChem-Substance/87226395
- PubChem-Substance/87226496
- PubChem-Substance/87226497
- PubChem-Substance/87226498
- PubChem-Substance/87226499
- PubChem-Substance/87226484
- PubChem-Substance/87226485
- PubChem-Substance/87226486
- PubChem-Substance/87226487
- PubChem-Substance/87226488
- PubChem-Substance/87226492
- PubChem-Substance/87226489
- PubChem-Substance/87226500
- PubChem-Substance/87226490
- PubChem-Substance/87226501
- PubChem-Substance/87226491
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Chemical Information
Molecular Biology Databases

