Automated detection of conformational epitopes using phage display Peptide sequences
- PMID: 20140073
- PMCID: PMC2808184
- DOI: 10.4137/bbi.s2745
Automated detection of conformational epitopes using phage display Peptide sequences
Abstract
Background: Precise determination of conformational epitopes of neutralizing antibodies represents a key step in the rational design of novel vaccines. A powerful experimental method to gain insights on the physical chemical nature of conformational epitopes is the selection of linear peptides that bind with high affinities to a monoclonal antibody of interest by phage display technology. However, the structural characterization of conformational epitopes from these mimotopes is not straightforward, and in the past the interpretation of peptide sequences from phage display experiments focused on linear sequence analysis to find a consensus sequence or common sequence motifs.
Results: We present a fully automated search method, EpiSearch that predicts the possible location of conformational epitopes on the surface of an antigen. The algorithm uses peptide sequences from phage display experiments as input, and ranks all surface exposed patches according to the frequency distribution of similar residues in the peptides and in the patch. We have tested the performance of the EpiSearch algorithm for six experimental data sets of phage display experiments, the human epidermal growth factor receptor-2 (HER-2/neu), the antibody mAb Bo2C11 targeting the C(2) domain of FVIII, antibodies mAb 17b and mAb b12 of the HIV envelope protein gp120, mAb 13b5 targeting HIV-1 capsid protein and 80R of the SARS coronavirus spike protein. In all these examples the conformational epitopes as determined by the X-ray crystal structures of the antibody-antigen complexes, were found within the highest scoring patches of EpiSearch, covering in most cases more than 50% residues of experimental observed conformational epitopes. Input options of the program include mapping of a single peptide or a set of peptides on the antigen structure, and the results of the calculation can be visualized on our interactive web server.
Availability: Users can access the EpiSearch from our web server http://curie.utmb.edu/episearch.html.
Keywords: conformational epitopes; peptides; phage display.
Figures

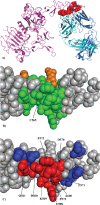
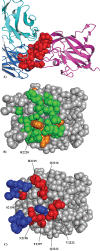
References
-
- Mirza O, Henriksen A, Ipsen H, et al. Dominant Epitopes and Allergic Cross-Reactivity: Complex Formation Between a Fab Fragment of a Monoclonal Murine IgG Antibody and the Major Allergen from Birch Pollen Bet v 1. J Immunol. 2000;165:331–8. - PubMed
-
- Padavattan S, Schirmer T, Schmidt M, et al. Identification of a B-cell Epitope of Hyaluronidase, a Major Bee Venom Allergen, from its Crystal Structure in Complex with a Specific Fab. Journal of Molecular Biology. 2007;368:742–52. - PubMed
-
- Padlan EA. X-ray crystallography of antibodies. Adv Protein Chem. 1996;49:57–133. - PubMed
-
- Spangfort MD, Mirza O, Ipsen H, van Neerven RJJ, Gajhede M, Larsen JN. Dominating IgE-Binding Epitope of Bet v 1, the Major Allergen of Birch Pollen, Characterized by X-ray Crystallography and Site-Directed Mutagenesis. J Immunol. 2003;171:3084–90. - PubMed
-
- Li Y, Li H, Yang F, Smith-Gill SJ, Mariuzza RA. X-ray snapshots of the maturation of an antibody response to a protein antigen. Nat Struct Mol Biol. 2003;10:482–8. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

