Deletion of the huntingtin polyglutamine stretch enhances neuronal autophagy and longevity in mice
- PMID: 20140187
- PMCID: PMC2816686
- DOI: 10.1371/journal.pgen.1000838
Deletion of the huntingtin polyglutamine stretch enhances neuronal autophagy and longevity in mice
Abstract
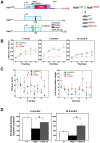
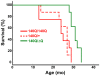
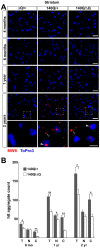
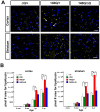
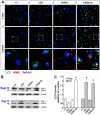
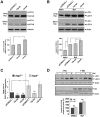
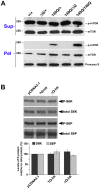
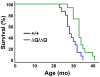
Expansion of a stretch of polyglutamine in huntingtin (htt), the protein product of the IT15 gene, causes Huntington's disease (HD). Previous investigations into the role of the polyglutamine stretch (polyQ) in htt function have suggested that its length may modulate a normal htt function involved in regulating energy homeostasis. Here we show that expression of full-length htt lacking its polyglutamine stretch (DeltaQ-htt) in a knockin mouse model for HD (Hdh(140Q/DeltaQ)), reduces significantly neuropil mutant htt aggregates, ameliorates motor/behavioral deficits, and extends lifespan in comparison to the HD model mice (Hdh(140Q/+)). The rescue of HD model phenotypes is accompanied by the normalization of lipofuscin levels in the brain and an increase in the steady-state levels of the mammalian autophagy marker microtubule-associate protein 1 light chain 3-II (LC3-II). We also find that DeltaQ-htt expression in vitro increases autophagosome synthesis and stimulates the Atg5-dependent clearance of truncated N-terminal htt aggregates. DeltaQ-htt's effect on autophagy most likely represents a gain-of-function, as overexpression of full-length wild-type htt in vitro does not increase autophagosome synthesis. Moreover, Hdh(DeltaQ/DeltaQ) mice live significantly longer than wild-type mice, suggesting that autophagy upregulation may be beneficial both in diseases caused by toxic intracellular aggregate-prone proteins and also as a lifespan extender in normal mammals.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
References
-
- Atwal RS, Xia J, Pinchev D, Taylor J, Epand RM, et al. Huntingtin has a membrane association signal that can modulate huntingtin aggregation, nuclear entry and toxicity. Hum Mol Genet. 2007;16:2600–2615. - PubMed
-
- Faber PW, Barnes GT, Srinidhi J, Chen J, Gusella JF, et al. Huntingtin interacts with a family of WW domain proteins. Hum Mol Genet. 1998;7:1463–1474. - PubMed
-
- Gao YG, Yan XZ, Song AX, Chang YG, Gao XC, et al. Structural Insights into the specific binding of huntingtin proline-rich region with the SH3 and WW domains. Structure. 2006;14:1755–1765. - PubMed
-
- Liu YF, Deth RC, Devys D. SH3 domain-dependent association of huntingtin with epidermal growth factor receptor signaling complexes. J Biol Chem. 1997;272:8121–8124. - PubMed
-
- The Huntington's Disease Collaborative Research Group. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. Cell. 1993;72:971–983. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

