The overlap of small molecule and protein binding sites within families of protein structures
- PMID: 20140189
- PMCID: PMC2816688
- DOI: 10.1371/journal.pcbi.1000668
The overlap of small molecule and protein binding sites within families of protein structures
Abstract
Protein-protein interactions are challenging targets for modulation by small molecules. Here, we propose an approach that harnesses the increasing structural coverage of protein complexes to identify small molecules that may target protein interactions. Specifically, we identify ligand and protein binding sites that overlap upon alignment of homologous proteins. Of the 2,619 protein structure families observed to bind proteins, 1,028 also bind small molecules (250-1000 Da), and 197 exhibit a statistically significant (p<0.01) overlap between ligand and protein binding positions. These "bi-functional positions", which bind both ligands and proteins, are particularly enriched in tyrosine and tryptophan residues, similar to "energetic hotspots" described previously, and are significantly less conserved than mono-functional and solvent exposed positions. Homology transfer identifies ligands whose binding sites overlap at least 20% of the protein interface for 35% of domain-domain and 45% of domain-peptide mediated interactions. The analysis recovered known small-molecule modulators of protein interactions as well as predicted new interaction targets based on the sequence similarity of ligand binding sites. We illustrate the predictive utility of the method by suggesting structural mechanisms for the effects of sanglifehrin A on HIV virion production, bepridil on the cellular entry of anthrax edema factor, and fusicoccin on vertebrate developmental pathways. The results, available at http://pibase.janelia.org, represent a comprehensive collection of structurally characterized modulators of protein interactions, and suggest that homologous structures are a useful resource for the rational design of interaction modulators.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
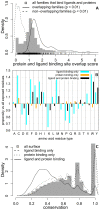
 0.01; solid; n = 197) or non-overlap (p
0.01; solid; n = 197) or non-overlap (p 0.01; dashed; n = 113). The highest overlap score observed is 10.83 (not shown). (B) The residue type propensity (Eqn 3) and (C) conservation (Eqn 4) at alignment positions that bind both ligands and proteins (black; n = 102,436), bind ligands (cyan; n = 46,610), bind proteins (orange; n = 491,723) in comparison to all solvent-exposed residues (grey; n = 1,147,882). The statistical significance of the residue propensities was estimated by a bootstrap resampling procedure (Table S5).
0.01; dashed; n = 113). The highest overlap score observed is 10.83 (not shown). (B) The residue type propensity (Eqn 3) and (C) conservation (Eqn 4) at alignment positions that bind both ligands and proteins (black; n = 102,436), bind ligands (cyan; n = 46,610), bind proteins (orange; n = 491,723) in comparison to all solvent-exposed residues (grey; n = 1,147,882). The statistical significance of the residue propensities was estimated by a bootstrap resampling procedure (Table S5).
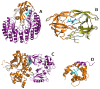
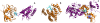
 (orange, 1A38:A) binding site for phosphopeptides (purple, 1A38:P). (C) Bepridil (cyan and blue, 1LXF:BEP) binds to Troponin C (grey, 1LXF:C) at positions that are homologous to the calmodulin (orange, 1K93:D) interface for anthrax edema factor (purple, 1K93:A). Troponin C aligns to both EF-hand motifs in calmodulin: The binding site aligned with EF-motif 2 (cyan) exhibits greater overlap with the anthrax edema factor interface than EF-motif 1 (blue).
(orange, 1A38:A) binding site for phosphopeptides (purple, 1A38:P). (C) Bepridil (cyan and blue, 1LXF:BEP) binds to Troponin C (grey, 1LXF:C) at positions that are homologous to the calmodulin (orange, 1K93:D) interface for anthrax edema factor (purple, 1K93:A). Troponin C aligns to both EF-hand motifs in calmodulin: The binding site aligned with EF-motif 2 (cyan) exhibits greater overlap with the anthrax edema factor interface than EF-motif 1 (blue).Similar articles
-
Proteome-wide prediction of overlapping small molecule and protein binding sites using structure.Mol Biosyst. 2011 Feb;7(2):545-57. doi: 10.1039/c0mb00200c. Epub 2010 Nov 24. Mol Biosyst. 2011. PMID: 21103609
-
Domain-based small molecule binding site annotation.BMC Bioinformatics. 2006 Mar 17;7:152. doi: 10.1186/1471-2105-7-152. BMC Bioinformatics. 2006. PMID: 16545112 Free PMC article.
-
Modulating protein-protein interactions with small molecules: the importance of binding hotspots.J Mol Biol. 2012 Jan 13;415(2):443-53. doi: 10.1016/j.jmb.2011.12.026. Epub 2011 Dec 16. J Mol Biol. 2012. PMID: 22198293 Free PMC article.
-
Challenges for the prediction of macromolecular interactions.Curr Opin Struct Biol. 2011 Jun;21(3):382-90. doi: 10.1016/j.sbi.2011.03.013. Epub 2011 Apr 14. Curr Opin Struct Biol. 2011. PMID: 21497504 Review.
-
Targeting protein-protein interactions with small molecules: challenges and perspectives for computational binding epitope detection and ligand finding.Curr Med Chem. 2006;13(22):2607-25. doi: 10.2174/092986706778201530. Curr Med Chem. 2006. PMID: 17017914 Review.
Cited by
-
PPDMs-a resource for mapping small molecule bioactivities from ChEMBL to Pfam-A protein domains.Bioinformatics. 2015 Mar 1;31(5):776-8. doi: 10.1093/bioinformatics/btu711. Epub 2014 Oct 27. Bioinformatics. 2015. PMID: 25348214 Free PMC article.
-
Drug-Like Protein-Protein Interaction Modulators: Challenges and Opportunities for Drug Discovery and Chemical Biology.Mol Inform. 2014 Jun;33(6-7):414-437. doi: 10.1002/minf.201400040. Epub 2014 Jun 2. Mol Inform. 2014. PMID: 25254076 Free PMC article. Review.
-
The protein interactome of Escherichia coli carbohydrate metabolism.PLoS One. 2025 Feb 4;20(2):e0315240. doi: 10.1371/journal.pone.0315240. eCollection 2025. PLoS One. 2025. PMID: 39903745 Free PMC article.
-
PoSSuM: a database of similar protein-ligand binding and putative pockets.Nucleic Acids Res. 2012 Jan;40(Database issue):D541-8. doi: 10.1093/nar/gkr1130. Epub 2011 Dec 1. Nucleic Acids Res. 2012. PMID: 22135290 Free PMC article.
-
Composition of Overlapping Protein-Protein and Protein-Ligand Interfaces.PLoS One. 2015 Oct 30;10(10):e0140965. doi: 10.1371/journal.pone.0140965. eCollection 2015. PLoS One. 2015. PMID: 26517868 Free PMC article.
References
-
- Berg T. Modulation of protein-protein interactions with small organic molecules. Angew Chem Int Ed Engl. 2003;42:2462–2481. - PubMed
-
- Wells JA, McClendon CL. Reaching for high-hanging fruit in drug discovery at protein-protein interfaces. Nature. 2007;450:1001–1009. - PubMed
-
- Arkin MR, Wells JA. Small-molecule inhibitors of protein-protein interactions: progressing towards the dream. Nat Rev Drug Discov. 2004;3:301–317. - PubMed
-
- Conte LL, Chothia C, Janin J. The atomic structure of protein-protein recognition sites. J Mol Biol. 1999;285:2177–2198. - PubMed

