A universal trend of reduced mRNA stability near the translation-initiation site in prokaryotes and eukaryotes
- PMID: 20140241
- PMCID: PMC2816680
- DOI: 10.1371/journal.pcbi.1000664
A universal trend of reduced mRNA stability near the translation-initiation site in prokaryotes and eukaryotes
Abstract
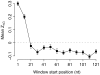
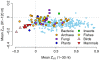
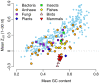
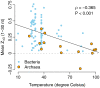
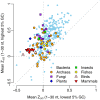
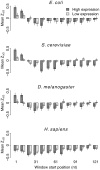
Recent studies have suggested that the thermodynamic stability of mRNA secondary structure near the start codon can regulate translation efficiency in Escherichia coli, and that translation is more efficient the less stable the secondary structure. We survey the complete genomes of 340 species for signals of reduced mRNA secondary structure near the start codon. Our analysis includes bacteria, archaea, fungi, plants, insects, fishes, birds, and mammals. We find that nearly all species show evidence for reduced mRNA stability near the start codon. The reduction in stability generally increases with increasing genomic GC content. In prokaryotes, the reduction also increases with decreasing optimal growth temperature. Within genomes, there is variation in the stability among genes, and this variation correlates with gene GC content, codon bias, and gene expression level. For birds and mammals, however, we do not find a genome-wide trend of reduced mRNA stability near the start codon. Yet the most GC rich genes in these organisms do show such a signal. We conclude that reduced stability of the mRNA secondary structure near the start codon is a universal feature of all cellular life. We suggest that the origin of this reduction is selection for efficient recognition of the start codon by initiator-tRNA.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Similar articles
-
Reduced stability of mRNA secondary structure near the translation-initiation site in dsDNA viruses.BMC Evol Biol. 2011 Mar 7;11:59. doi: 10.1186/1471-2148-11-59. BMC Evol Biol. 2011. PMID: 21385374 Free PMC article.
-
How Many Messenger RNAs Can Be Translated by the START Mechanism?Int J Mol Sci. 2020 Nov 8;21(21):8373. doi: 10.3390/ijms21218373. Int J Mol Sci. 2020. PMID: 33171614 Free PMC article.
-
Initiator AUGs Are Discriminated from Elongator AUGs Predominantly through mRNA Accessibility in C. crescentus.J Bacteriol. 2023 May 25;205(5):e0042022. doi: 10.1128/jb.00420-22. Epub 2023 Apr 24. J Bacteriol. 2023. PMID: 37092987 Free PMC article.
-
The interplay between cis- and trans-acting factors drives selective mRNA translation initiation in eukaryotes.Biochimie. 2024 Feb;217:20-30. doi: 10.1016/j.biochi.2023.09.017. Epub 2023 Sep 21. Biochimie. 2024. PMID: 37741547 Review.
-
AUGcontext DB: a comprehensive catalog of the mRNA AUG initiator codon context across eukaryotes.RNA Biol. 2025 Dec;22(1):1-5. doi: 10.1080/15476286.2025.2465196. Epub 2025 Feb 13. RNA Biol. 2025. PMID: 39936323 Free PMC article. Review.
Cited by
-
Decoding mechanisms by which silent codon changes influence protein biogenesis and function.Int J Biochem Cell Biol. 2015 Jul;64:58-74. doi: 10.1016/j.biocel.2015.03.011. Epub 2015 Mar 26. Int J Biochem Cell Biol. 2015. PMID: 25817479 Free PMC article. Review.
-
Number variation of high stability regions is correlated with gene functions.Genome Biol Evol. 2013;5(3):484-93. doi: 10.1093/gbe/evt020. Genome Biol Evol. 2013. PMID: 23407773 Free PMC article.
-
Non-optimal codon usage is a mechanism to achieve circadian clock conditionality.Nature. 2013 Mar 7;495(7439):116-20. doi: 10.1038/nature11942. Epub 2013 Feb 17. Nature. 2013. PMID: 23417065 Free PMC article.
-
Accounting for experimental noise reveals that mRNA levels, amplified by post-transcriptional processes, largely determine steady-state protein levels in yeast.PLoS Genet. 2015 May 7;11(5):e1005206. doi: 10.1371/journal.pgen.1005206. eCollection 2015 May. PLoS Genet. 2015. PMID: 25950722 Free PMC article.
-
Nucleotide sequence composition adjacent to intronic splice sites improves splicing efficiency via its effect on pre-mRNA local folding in fungi.RNA. 2015 Oct;21(10):1704-18. doi: 10.1261/rna.051268.115. Epub 2015 Aug 5. RNA. 2015. PMID: 26246046 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

