Primary metabolism of chickpea is the initial target of wound inducing early sensed Fusarium oxysporum f. sp. ciceri race I
- PMID: 20140256
- PMCID: PMC2815786
- DOI: 10.1371/journal.pone.0009030
Primary metabolism of chickpea is the initial target of wound inducing early sensed Fusarium oxysporum f. sp. ciceri race I
Abstract
Background: Biotrophic interaction between host and pathogen induces generation of reactive oxygen species that leads to programmed cell death of the host tissue specifically encompassing the site of infection conferring resistance to the host. However, in the present study, biotrophic relationship between Fusarium oxysporum and chickpea provided some novel insights into the classical concepts of defense signaling and disease perception where ROS (reactive oxygen species) generation followed by hypersensitive responses determined the magnitude of susceptibility or resistant potentiality of the host.
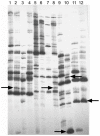
Methodology/principal findings: Microscopic observations detected wound mediated in planta pathogenic establishment and its gradual progression within the host vascular tissue. cDNA-AFLP showed differential expression of many defense responsive elements. Real time expression profiling also validated the early recognition of the wound inducing pathogen by the host. The interplay between fungus and host activated changes in primary metabolism, which generated defense signals in the form of sugar molecules for combating pathogenic encounter.
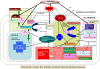
Conclusions/significance: The present study showed the limitations of hypersensitive response mediated resistance, especially when foreign encounters involved the food production as well as the translocation machinery of the host. It was also predicted from the obtained results that hypersensitivity and active species generation failed to impart host defense in compatible interaction between chickpea and Fusarium. On the contrary, the defense related gene(s) played a critical role in conferring natural resistance to the resistant host. Thus, this study suggests that natural selection is the decisive factor for selecting and segregating out the suitable type of defense mechanism to be undertaken by the host without disturbing its normal metabolism, which could deviate from the known classical defense mechanisms.
Conflict of interest statement
Figures




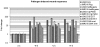


References
-
- Dixon RA, Harrison MJ, Lamb CJ. Early events in the activation of plant defense responses. Ann Rev Phytopathol. 1994;32:479–501.
-
- Glazebrook J. Contrasting mechanism of defense against biotrophic and necrotrophic pathogens. Ann Rev Phytopathol. 2005;43:205–227. - PubMed
-
- Doehlemann G, Ramon W, Horst RJ, Voll LM, Usadel B, et al. Reprogramming a maize plant: transcriptional and metabolic changes induced by the fungal biotroph Ustilago maydis. Plant J. 2008;56:181–195. - PubMed
-
- Mendgen K, Hahn M. Plant infection and the establishment of fungal biotrophy. Trends Plant Sci. 2002;7:352–356. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

