Contribution of genome-wide HCV genetic differences to outcome of interferon-based therapy in Caucasian American and African American patients
- PMID: 20140258
- PMCID: PMC2815788
- DOI: 10.1371/journal.pone.0009032
Contribution of genome-wide HCV genetic differences to outcome of interferon-based therapy in Caucasian American and African American patients
Abstract
Background: Hepatitis C virus (HCV) has six major genotypes, and patients infected with genotype 1 respond less well to interferon-based therapy than other genotypes. African American patients respond to interferon alpha-based therapy at about half the rate of Caucasian Americans. The effect of HCV's genetic variation on treatment outcome in both racial groups is poorly understood.
Methodology: We determined the near full-length pre-therapy consensus sequences from 94 patients infected with HCV genotype 1a or 1b undergoing treatment with peginterferon alpha-2a and ribavirin through the Virahep-C study. The sequences were stratified by genotype, race and treatment outcome to identify HCV genetic differences associated with treatment efficacy.
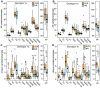
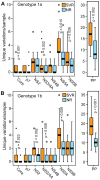
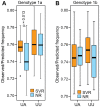
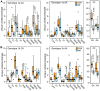
Principal findings: HCV sequences from patients who achieved sustained viral response were more diverse than sequences from non-responders. These inter-patient diversity differences were found primarily in the NS5A gene in genotype 1a and in core and NS2 in genotype 1b. These differences could not be explained by host selection pressures. Genotype 1b but not 1a African American patients had viral genetic differences that correlated with treatment outcome.
Conclusions & significance: Higher inter-patient viral genetic diversity correlated with successful treatment, implying that there are HCV genotype 1 strains with intrinsic differences in sensitivity to therapy. Core, NS3 and NS5A have interferon-suppressive activities detectable through in vitro assays, and hence these activities also appear to function in human patients. Both preferential infection with relatively resistant HCV variants and host-specific factors appear to contribute to the unusually poor response to therapy in African American patients.
Conflict of interest statement
Figures


Similar articles
-
Pretreatment sequence diversity differences in the full-length hepatitis C virus open reading frame correlate with early response to therapy.J Virol. 2007 Aug;81(15):8211-24. doi: 10.1128/JVI.00487-07. Epub 2007 May 23. J Virol. 2007. PMID: 17522222 Free PMC article. Clinical Trial.
-
Quasispecies of hepatitis C virus genotype 1 and treatment outcome with peginterferon and ribavirin.Infect Genet Evol. 2009 Jul;9(4):689-98. doi: 10.1016/j.meegid.2008.11.001. Epub 2008 Nov 21. Infect Genet Evol. 2009. PMID: 19063998
-
Impact of NS5A sequences of Hepatitis C virus genotype 1a on early viral kinetics during treatment with peginterferon- alpha 2a plus ribavirin.J Infect Dis. 2007 Oct 1;196(7):998-1005. doi: 10.1086/521306. Epub 2007 Aug 21. J Infect Dis. 2007. PMID: 17763320 Clinical Trial.
-
Importance of HCV genotype 1 subtypes for drug resistance and response to therapy.J Viral Hepat. 2014 Apr;21(4):229-40. doi: 10.1111/jvh.12230. J Viral Hepat. 2014. PMID: 24597691 Review.
-
Viral kinetics in hepatitis C virus: special patient populations.Semin Liver Dis. 2003;23 Suppl 1:29-33. doi: 10.1055/s-2003-41632. Semin Liver Dis. 2003. PMID: 12934166 Review.
Cited by
-
Assessment of hepatitis C virus protein sequences with regard to interferon/ribavirin combination therapy response in patients with HCV genotype 1b.Protein J. 2012 Feb;31(2):129-36. doi: 10.1007/s10930-011-9381-6. Protein J. 2012. PMID: 22170451
-
Analysis of the complete open reading frame of hepatitis C virus in genotype 2a infection reveals critical sites influencing the response to peginterferon and ribavirin therapy.Hepatol Int. 2011 Sep;5(3):789-99. doi: 10.1007/s12072-011-9267-x. Epub 2011 Mar 20. Hepatol Int. 2011. PMID: 21484117
-
Identification and comparative analysis of hepatitis C virus-host cell protein interactions.Mol Biosyst. 2013 Dec;9(12):3199-209. doi: 10.1039/c3mb70343f. Epub 2013 Oct 18. Mol Biosyst. 2013. PMID: 24136289 Free PMC article.
-
Analysis of the complete open reading frame of genotype 2b hepatitis C virus in association with the response to peginterferon and ribavirin therapy.PLoS One. 2011;6(9):e24514. doi: 10.1371/journal.pone.0024514. Epub 2011 Sep 15. PLoS One. 2011. PMID: 21935415 Free PMC article.
-
Disparities and risks of sexually transmissible infections among men who have sex with men in China: a meta-analysis and data synthesis.PLoS One. 2014 Feb 24;9(2):e89959. doi: 10.1371/journal.pone.0089959. eCollection 2014. PLoS One. 2014. PMID: 24587152 Free PMC article.
References
-
- Armstrong GL, Wasley A, Simard EP, McQuillan GM, Kuhnert WL, et al. The prevalence of hepatitis C virus infection in the United States, 1999 through 2002. Ann Intern Med. 2006;144:705–714. - PubMed
-
- McHutchison JG, Bacon BR. Chronic hepatitis C: an age wave of disease burden. Am J Manag Care. 2005;11:S286–S295. - PubMed
-
- Lemon SM, Walker C, Alter MJ, Yi M. Hepatitis C Virus. In: Knipe DM, Howley P, Griffin DE, Lamb RA, Martin MA, editors. Fields Virology. Philadelphia, PA: Lippicott, Williams & Wilkins; 2007. pp. 1253–1304.
-
- Smith DB, Pathirana S, Davidson F, Lawlor E, Power J, et al. The origin of hepatitis C virus genotypes. J Gen Virol. 1997;78 (Pt2):321–328. - PubMed
-
- Fried MW, Shiffman ML, Reddy KR, Smith C, Marinos G, et al. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N Engl J Med. 2002;347:975–982. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

