Ligand-free open-closed transitions of periplasmic binding proteins: the case of glutamine-binding protein
- PMID: 20141110
- PMCID: PMC2831130
- DOI: 10.1021/bi902045p
Ligand-free open-closed transitions of periplasmic binding proteins: the case of glutamine-binding protein
Abstract
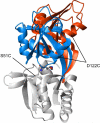
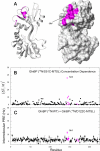
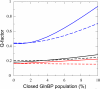
The ability to undergo large-scale domain rearrangements is essential for the substrate-binding function of periplasmic binding proteins (PBPs), which are indispensable for nutrient uptake in Gram-negative bacteria. Crystal structures indicate that PBPs typically adopt either an "open" unliganded configuration or a "closed" liganded one. However, it is not clear whether, as a general rule, PBPs remain open until ligand-induced interdomain closure or are in equilibrium with a minor population of unliganded, closed species. Evidence for the latter has been recently reported on maltose-binding protein (MBP) in aqueous solution [Tang, C., et al. (2007) Nature 449, 1078-1082] via paramagnetic relaxation enhancement (PRE), a technique able to probe lowly populated regions of conformational space. Here, we use PRE to study the unliganded open-closed transition of another PBP: glutamine-binding protein (GlnBP). Through a combination of domain structure knowledge and intermolecular and concentration dependence PRE experiments, a set of surface residues was found to be involved in intermolecular interactions. Barring such residues, PRE data on ligand-free GlnBP, paramagnetically labeled at two sites (one at a time), could be appropriately explained by the unliganded, open crystal structure in that it both yielded a good PRE fit and was not significantly affected by PRE-based refinement. Thus, contrary to MBP, our data did not particularly suggest the coexistence of a minor closed conformer. Several possibilities were explored to explain the observed differences in such closely structurally related systems; among them, a particularly interesting one arises from close inspection of the interdomain "hinge" region of various PBPs: strong hydrogen bond interactions discourage large-scale interdomain dynamics.
Figures
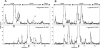


Similar articles
-
Accessing a hidden conformation of the maltose binding protein using accelerated molecular dynamics.PLoS Comput Biol. 2011 Apr;7(4):e1002034. doi: 10.1371/journal.pcbi.1002034. Epub 2011 Apr 21. PLoS Comput Biol. 2011. PMID: 21533070 Free PMC article.
-
Trapping open and closed forms of FitE: a group III periplasmic binding protein.Proteins. 2009 May 15;75(3):598-609. doi: 10.1002/prot.22272. Proteins. 2009. PMID: 19004000
-
Full engagement of liganded maltose-binding protein stabilizes a semi-open ATP-binding cassette dimer in the maltose transporter.Mol Microbiol. 2015 Dec;98(5):878-94. doi: 10.1111/mmi.13165. Epub 2015 Sep 10. Mol Microbiol. 2015. PMID: 26268698 Free PMC article.
-
The crystal structure of glutamine-binding protein from Escherichia coli.J Mol Biol. 1996 Sep 20;262(2):225-42. doi: 10.1006/jmbi.1996.0509. J Mol Biol. 1996. PMID: 8831790 Review.
-
Molecular mechanism of ferricsiderophore passage through the outer membrane receptor proteins of Escherichia coli.Biometals. 2007 Jun;20(3-4):263-74. doi: 10.1007/s10534-006-9060-9. Epub 2006 Dec 22. Biometals. 2007. PMID: 17186377 Review.
Cited by
-
Exploration of multi-state conformational dynamics and underlying global functional landscape of maltose binding protein.PLoS Comput Biol. 2012;8(4):e1002471. doi: 10.1371/journal.pcbi.1002471. Epub 2012 Apr 19. PLoS Comput Biol. 2012. PMID: 22532792 Free PMC article.
-
A single-molecule dissection of ligand binding to a protein with intrinsic dynamics.Nat Chem Biol. 2013 May;9(5):313-8. doi: 10.1038/nchembio.1213. Epub 2013 Mar 17. Nat Chem Biol. 2013. PMID: 23502425
-
NMR-based conformational ensembles explain pH-gated opening and closing of OmpG channel.J Am Chem Soc. 2013 Oct 9;135(40):15101-13. doi: 10.1021/ja408206e. Epub 2013 Oct 1. J Am Chem Soc. 2013. PMID: 24020969 Free PMC article.
-
Temperature dependence of molecular interactions involved in defining stability of glutamine binding protein and its complex with L-glutamine.Biochemistry. 2012 Jan 17;51(2):643-52. doi: 10.1021/bi201494h. Epub 2012 Jan 6. Biochemistry. 2012. PMID: 22206385 Free PMC article.
-
Accessing a hidden conformation of the maltose binding protein using accelerated molecular dynamics.PLoS Comput Biol. 2011 Apr;7(4):e1002034. doi: 10.1371/journal.pcbi.1002034. Epub 2011 Apr 21. PLoS Comput Biol. 2011. PMID: 21533070 Free PMC article.
References
-
- Doeven MK, Kok J, Poolman B. Specificity and selectivity determinants of peptide transport in Lactococcus lactis and other microorganisms. Molecular Microbiology. 2005;57:640–649. - PubMed
-
- Quiocho FA, Ledvina PS. Atomic structure and specificity of bacterial periplasmic receptors for active transport and chemotaxis: Variation of common themes. Molecular Microbiology. 1996;20:17–25. - PubMed
-
- Monod J, Wyman J, Changeux JP. On Nature of Allosteric Transitions - a Plausible Model. Journal of Molecular Biology. 1965;12:88–118. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

