New insights into the blood-stage transcriptome of Plasmodium falciparum using RNA-Seq
- PMID: 20141604
- PMCID: PMC2859250
- DOI: 10.1111/j.1365-2958.2009.07026.x
New insights into the blood-stage transcriptome of Plasmodium falciparum using RNA-Seq
Abstract
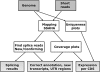
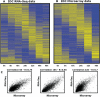
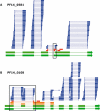
Recent advances in high-throughput sequencing present a new opportunity to deeply probe an organism's transcriptome. In this study, we used Illumina-based massively parallel sequencing to gain new insight into the transcriptome (RNA-Seq) of the human malaria parasite, Plasmodium falciparum. Using data collected at seven time points during the intraerythrocytic developmental cycle, we (i) detect novel gene transcripts; (ii) correct hundreds of gene models; (iii) propose alternative splicing events; and (iv) predict 5' and 3' untranslated regions. Approximately 70% of the unique sequencing reads map to previously annotated protein-coding genes. The RNA-Seq results greatly improve existing annotation of the P. falciparum genome with over 10% of gene models modified. Our data confirm 75% of predicted splice sites and identify 202 new splice sites, including 84 previously uncharacterized alternative splicing events. We also discovered 107 novel transcripts and expression of 38 pseudogenes, with many demonstrating differential expression across the developmental time series. Our RNA-Seq results correlate well with DNA microarray analysis performed in parallel on the same samples, and provide improved resolution over the microarray-based method. These data reveal new features of the P. falciparum transcriptional landscape and significantly advance our understanding of the parasite's red blood cell-stage transcriptome.
Figures


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

