Variability of gene expression profiles in human blood and lymphoblastoid cell lines
- PMID: 20141636
- PMCID: PMC2841682
- DOI: 10.1186/1471-2164-11-96
Variability of gene expression profiles in human blood and lymphoblastoid cell lines
Abstract
Background: Readily accessible samples such as peripheral blood or cell lines are increasingly being used in large cohorts to characterise gene expression differences between a patient group and healthy controls. However, cell and RNA isolation procedures and the variety of cell types that make up whole blood can affect gene expression measurements. We therefore systematically investigated global gene expression profiles in peripheral blood from six individuals collected during two visits by comparing five of the following cell and RNA isolation methods: whole blood (PAXgene), peripheral blood mononuclear cells (PBMCs), lymphoblastoid cell lines (LCLs), CD19 and CD20 specific B-cell subsets.
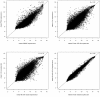
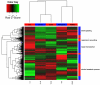
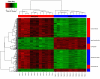
Results: Gene expression measurements were clearly discriminated by isolation method although the reproducibility was high for all methods (range rho = 0.90-1.00). The PAXgene samples showed a decrease in the number of expressed genes (P < 1*10(-16)) with higher variability (P < 1*10(-16)) compared to the other methods. Differentially expressed probes between PAXgene and PBMCs were correlated with the number of monocytes, lymphocytes, neutrophils or erythrocytes. The correlations (rho = 0.83; rho = 0.79) of the expression levels of detected probes between LCLs and B-cell subsets were much lower compared to the two B-cell isolation methods (rho = 0.98). Gene ontology analysis of detected genes showed that genes involved in inflammatory responses are enriched in B-cells CD19 and CD20 whereas genes involved in alcohol metabolic process and the cell cycle were enriched in LCLs.
Conclusion: Gene expression profiles in blood-based samples are strongly dependent on the predominant constituent cell type(s) and RNA isolation method. It is crucial to understand the differences and variability of gene expression measurements between cell and RNA isolation procedures, and their relevance to disease processes, before application in large clinical studies.
Figures


Similar articles
-
Differential gene expression profiles are dependent upon method of peripheral blood collection and RNA isolation.BMC Genomics. 2008 Oct 10;9:474. doi: 10.1186/1471-2164-9-474. BMC Genomics. 2008. PMID: 18847473 Free PMC article.
-
Investigating gene expression profiles of whole blood and peripheral blood mononuclear cells using multiple collection and processing methods.PLoS One. 2019 Dec 6;14(12):e0225137. doi: 10.1371/journal.pone.0225137. eCollection 2019. PLoS One. 2019. PMID: 31809517 Free PMC article.
-
RNA from stabilized whole blood enables more comprehensive immune gene expression profiling compared to RNA from peripheral blood mononuclear cells.PLoS One. 2020 Jun 26;15(6):e0235413. doi: 10.1371/journal.pone.0235413. eCollection 2020. PLoS One. 2020. PMID: 32589655 Free PMC article.
-
Whole blood and leukocyte RNA isolation for gene expression analyses.Physiol Genomics. 2004 Nov 17;19(3):247-54. doi: 10.1152/physiolgenomics.00020.2004. Physiol Genomics. 2004. PMID: 15548831
-
Gene expression profiling of whole blood: A comparative assessment of RNA-stabilizing collection methods.PLoS One. 2019 Oct 10;14(10):e0223065. doi: 10.1371/journal.pone.0223065. eCollection 2019. PLoS One. 2019. PMID: 31600258 Free PMC article.
Cited by
-
Transcriptomic analysis of circulating leukocytes reveals novel aspects of the host systemic inflammatory response to sheep scab mites.PLoS One. 2012;7(8):e42778. doi: 10.1371/journal.pone.0042778. Epub 2012 Aug 3. PLoS One. 2012. PMID: 22880105 Free PMC article.
-
Genetic control of gene expression in whole blood and lymphoblastoid cell lines is largely independent.Genome Res. 2012 Mar;22(3):456-66. doi: 10.1101/gr.126540.111. Epub 2011 Dec 19. Genome Res. 2012. PMID: 22183966 Free PMC article.
-
Functional analysis and molecular characterization of spontaneously outgrown human lymphoblastoid cell lines.Mol Biol Rep. 2014 Oct;41(10):6995-7007. doi: 10.1007/s11033-014-3587-6. Epub 2014 Jul 19. Mol Biol Rep. 2014. PMID: 25037273
-
Biomarkers of Nutrition and Health: New Tools for New Approaches.Nutrients. 2019 May 16;11(5):1092. doi: 10.3390/nu11051092. Nutrients. 2019. PMID: 31100942 Free PMC article. Review.
-
Blood-based identification of non-responders to anti-TNF therapy in rheumatoid arthritis.BMC Med Genomics. 2015 Jun 3;8:26. doi: 10.1186/s12920-015-0100-6. BMC Med Genomics. 2015. PMID: 26036272 Free PMC article.
References
-
- Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, Boldrick JC, Sabet H, Tran T, Yu X, Powell JI, Yang L, Marti GE, Moore T, Hudson J Jr, Lu L, Lewis DB, Tibshirani R, Sherlock G, Chan WC, Greiner TC, Weisenburger DD, Armitage JO, Warnke R, Levy R, Wilson W, Grever MR, Byrd JC, Botstein D, Brown PO, Staudt LM. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. - DOI - PubMed
-
- Khan J, Wei JS, Ringner M, Saal LH, Ladanyi M, Westermann F, Berthold F, Schwab M, Antonescu CR, Peterson C, Meltzer PS. Classification and diagnostic prediction of cancers using gene expression profiling and artificial neural networks. Nat Med. 2001;7:673–679. doi: 10.1038/89044. - DOI - PMC - PubMed
-
- Vijver MJ van de, He YD, van't Veer LJ, Dai H, Hart AA, Voskuil DW, Schreiber GJ, Peterse JL, Roberts C, Marton MJ, Parrish M, Atsma D, Witteveen A, Glas A, Delahaye L, van d V, Bartelink H, Rodenhuis S, Rutgers ET, Friend SH, Bernards R. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

