The histone deacetylase Sirt6 regulates glucose homeostasis via Hif1alpha
- PMID: 20141841
- PMCID: PMC2821045
- DOI: 10.1016/j.cell.2009.12.041
The histone deacetylase Sirt6 regulates glucose homeostasis via Hif1alpha
Abstract
SIRT6 is a member of a highly conserved family of NAD(+)-dependent deacetylases with various roles in metabolism, stress resistance, and life span. SIRT6-deficient mice develop normally but succumb to a lethal hypoglycemia early in life; however, the mechanism underlying this hypoglycemia remained unclear. Here, we demonstrate that SIRT6 functions as a histone H3K9 deacetylase to control the expression of multiple glycolytic genes. Specifically, SIRT6 appears to function as a corepressor of the transcription factor Hif1alpha, a critical regulator of nutrient stress responses. Consistent with this notion, SIRT6-deficient cells exhibit increased Hif1alpha activity and show increased glucose uptake with upregulation of glycolysis and diminished mitochondrial respiration. Our studies uncover a role for the chromatin factor SIRT6 as a master regulator of glucose homeostasis and may provide the basis for novel therapeutic approaches against metabolic diseases, such as diabetes and obesity.
Copyright 2010 Elsevier Inc. All rights reserved.
Figures


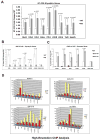



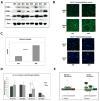
References
-
- Aragones J, Fraisl P, Baes M, Carmeliet P. Oxygen sensors at the crossroad of metabolism. Cell metabolism. 2009;9:11–22. - PubMed
-
- Bassing CH, Chua KF, Sekiguchi J, Suh H, Whitlow SR, Fleming JC, Monroe BC, Ciccone DN, Yan C, Vlasakova K, et al. Increased ionizing radiation sensitivity and genomic instability in the absence of histone H2AX. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:8173–8178. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

