Comparative analyses of seven algorithms for copy number variant identification from single nucleotide polymorphism arrays
- PMID: 20142258
- PMCID: PMC2875020
- DOI: 10.1093/nar/gkq040
Comparative analyses of seven algorithms for copy number variant identification from single nucleotide polymorphism arrays
Abstract
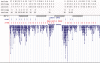
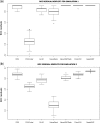
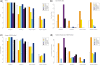
Determination of copy number variants (CNVs) inferred in genome wide single nucleotide polymorphism arrays has shown increasing utility in genetic variant disease associations. Several CNV detection methods are available, but differences in CNV call thresholds and characteristics exist. We evaluated the relative performance of seven methods: circular binary segmentation, CNVFinder, cnvPartition, gain and loss of DNA, Nexus algorithms, PennCNV and QuantiSNP. Tested data included real and simulated Illumina HumHap 550 data from the Singapore cohort study of the risk factors for Myopia (SCORM) and simulated data from Affymetrix 6.0 and platform-independent distributions. The normalized singleton ratio (NSR) is proposed as a metric for parameter optimization before enacting full analysis. We used 10 SCORM samples for optimizing parameter settings for each method and then evaluated method performance at optimal parameters using 100 SCORM samples. The statistical power, false positive rates, and receiver operating characteristic (ROC) curve residuals were evaluated by simulation studies. Optimal parameters, as determined by NSR and ROC curve residuals, were consistent across datasets. QuantiSNP outperformed other methods based on ROC curve residuals over most datasets. Nexus Rank and SNPRank have low specificity and high power. Nexus Rank calls oversized CNVs. PennCNV detects one of the fewest numbers of CNVs.
Figures

References
-
- Feuk L, Carson AR, Scherer SW. Structural variation in the human genome. Nat. Rev. Genet. 2006;7:85–97. - PubMed
-
- Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, Qi Y, Scherer SW, Lee C. Detection of large-scale variation in the human genome. Nat. Genet. 2004;36:949–951. - PubMed
-
- Jakobsson M, Scholz SW, Scheet P, Gibbs JR, VanLiere JM, Fung HC, Szpiech ZA, Degnan JH, Wang K, Guerreiro R, et al. Genotype, haplotype and copy-number variation in worldwide human populations. Nature. 2008;451:998–1003. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

