Cross-species analysis traces adaptation of Rubisco toward optimality in a low-dimensional landscape
- PMID: 20142476
- PMCID: PMC2840432
- DOI: 10.1073/pnas.0911663107
Cross-species analysis traces adaptation of Rubisco toward optimality in a low-dimensional landscape
Abstract
Rubisco (D-ribulose 1,5-bisphosphate carboxylase/oxygenase), probably the most abundant protein in the biosphere, performs an essential part in the process of carbon fixation through photosynthesis, thus facilitating life on earth. Despite the significant effect that Rubisco has on the fitness of plants and other photosynthetic organisms, this enzyme is known to have a low catalytic rate and a tendency to confuse its substrate, carbon dioxide, with oxygen. This apparent inefficiency is puzzling and raises questions regarding the roles of evolution versus biochemical constraints in shaping Rubisco. Here we examine these questions by analyzing the measured kinetic parameters of Rubisco from various organisms living in various environments. The analysis presented here suggests that the evolution of Rubisco is confined to an effectively one-dimensional landscape, which is manifested in simple power law correlations between its kinetic parameters. Within this one-dimensional landscape, which may represent biochemical and structural constraints, Rubisco appears to be tuned to the intracellular environment in which it resides such that the net photosynthesis rate is nearly optimal. Our analysis indicates that the specificity of Rubisco is not the main determinant of its efficiency but rather the trade-off between the carboxylation velocity and CO(2) affinity. As a result, the presence of oxygen has only a moderate effect on the optimal performance of Rubisco, which is determined mostly by the local CO(2) concentration. Rubisco appears as an experimentally testable example for the evolution of proteins subject both to strong selection pressure and to biochemical constraints that strongly confine the evolutionary plasticity to a low-dimensional landscape.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


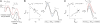
 ) decrease in the O2 addition barrier, ΔG
1,O, such that 0.5 · ΔG
1,C – ΔG
2,O = const. The negative correlation between v
C and S is the outcome of the two trade-offs: As v
C decreases, k
on,C increases and, as a result, k
on,O also increases, but by a smaller factor, and the difference between the gas addition barriers increases resulting in a decrease of the specificity, S = exp(ΔG
1,O – ΔG
1,C).
) decrease in the O2 addition barrier, ΔG
1,O, such that 0.5 · ΔG
1,C – ΔG
2,O = const. The negative correlation between v
C and S is the outcome of the two trade-offs: As v
C decreases, k
on,C increases and, as a result, k
on,O also increases, but by a smaller factor, and the difference between the gas addition barriers increases resulting in a decrease of the specificity, S = exp(ΔG
1,O – ΔG
1,C).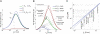
Similar articles
-
RubisCO Early Oxygenase Activity: A Kinetic and Evolutionary Perspective.Bioessays. 2017 Nov;39(11). doi: 10.1002/bies.201700071. Epub 2017 Oct 4. Bioessays. 2017. PMID: 28976010 Review.
-
Evolutionary trends in RuBisCO kinetics and their co-evolution with CO2 concentrating mechanisms.Plant J. 2020 Feb;101(4):897-918. doi: 10.1111/tpj.14643. Epub 2020 Jan 2. Plant J. 2020. PMID: 31820505 Review.
-
Environmentally driven evolution of Rubisco and improved photosynthesis and growth within the C3 genus Limonium (Plumbaginaceae).New Phytol. 2014 Aug;203(3):989-99. doi: 10.1111/nph.12858. Epub 2014 May 23. New Phytol. 2014. PMID: 24861241
-
Expanding knowledge of the Rubisco kinetics variability in plant species: environmental and evolutionary trends.Plant Cell Environ. 2014 Sep;37(9):1989-2001. doi: 10.1111/pce.12335. Epub 2014 May 11. Plant Cell Environ. 2014. PMID: 24689692
-
Modelling (18)O2 and (16)O2 unidirectional fluxes in plants. III: fitting of experimental data by a simple model.Biosystems. 2013 Aug;113(2):104-14. doi: 10.1016/j.biosystems.2012.10.004. Epub 2012 Nov 13. Biosystems. 2013. PMID: 23153764
Cited by
-
Structural mechanism of RuBisCO activation by carbamylation of the active site lysine.Proc Natl Acad Sci U S A. 2012 Nov 13;109(46):18785-90. doi: 10.1073/pnas.1210754109. Epub 2012 Oct 29. Proc Natl Acad Sci U S A. 2012. PMID: 23112176 Free PMC article.
-
Modeling genome-wide enzyme evolution predicts strong epistasis underlying catalytic turnover rates.Nat Commun. 2018 Dec 10;9(1):5270. doi: 10.1038/s41467-018-07649-1. Nat Commun. 2018. PMID: 30532008 Free PMC article.
-
Optimization of enzyme parameters for fermentative production of biorenewable fuels and chemicals.Comput Struct Biotechnol J. 2012 Oct 31;3:e201210005. doi: 10.5936/csbj.201210005. eCollection 2012. Comput Struct Biotechnol J. 2012. PMID: 24688665 Free PMC article. Review.
-
Temperature responses of the Rubisco maximum carboxylase activity across domains of life: phylogenetic signals, trade-offs, and importance for carbon gain.Photosynth Res. 2015 Feb;123(2):183-201. doi: 10.1007/s11120-014-0067-8. Epub 2014 Dec 17. Photosynth Res. 2015. PMID: 25515770
-
Photosynthesis research under climate change.Photosynth Res. 2021 Dec;150(1-3):5-19. doi: 10.1007/s11120-021-00861-z. Epub 2021 Jul 7. Photosynth Res. 2021. PMID: 34235625 Review.
References
-
- Ellis RJ. The most abundant protein in the world. Trends Biochem Sci. 1979;4:241–244.
-
- Andrews TJ, Lorimer GH. Photorespiration—Still unavoidable? FEBS Lett. 1978;90:1–9.
-
- Lorimer GH, Andrews TJ. Plant photorespiration—An inevitable consequence of the existence of atmospheric oxygen. Nature. 1973;243:359–360.
-
- Andersson I, Backlund A. Structure and function of Rubisco. Plant Physiol Biochem. 2008;46:275–291. - PubMed
-
- Bowsher C, Steer M, Tobin A. Plant Biochemistry. New York: Taylor & Francis; 2008.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

