Abasic sites and strand breaks in DNA cause transcriptional mutagenesis in Escherichia coli
- PMID: 20142484
- PMCID: PMC2840506
- DOI: 10.1073/pnas.0913191107
Abasic sites and strand breaks in DNA cause transcriptional mutagenesis in Escherichia coli
Abstract
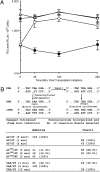
DNA damage occurs continuously, and faithful replication and transcription are essential for maintaining cell viability. Cells in nature are not dividing and replicating DNA often; therefore it is important to consider the outcome of RNA polymerase (RNAP) encounters with DNA damage. Base damage in the DNA can affect transcriptional fidelity, leading to production of mutant mRNA and protein in a process termed transcriptional mutagenesis (TM). Abasic (AP) sites and strand breaks are frequently occurring, spontaneous damages that are also base excision repair (BER) intermediates. In vitro studies have demonstrated that these lesions can be bypassed by RNAP; however this has never been assessed in vivo. This study demonstrates that RNAP is capable of bypassing AP sites and strand breaks in Escherichia coli and results in TM through adenine incorporation in nascent mRNA. Elimination of the enzymes that process these lesions further increases TM; however, such mutants can still complete repair by other downstream pathways. These results show that AP sites and strand breaks can result in mutagenic RNAP bypass and have important implications for the biologic endpoints of DNA damage.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Nouspikel T, Hanawalt PC. DNA repair in terminally differentiated cells. DNA Repair (Amst) 2002;1:59–75. - PubMed
-
- Savageau MA. Escherichia coli habitats, cell types, and molecular mechanisms of gene control. Am Nat. 1983;(122):732–744.
-
- Brégeon D, Doddridge ZA, You HJ, Weiss B, Doetsch PW. Transcriptional mutagenesis induced by uracil and 8-oxoguanine in Escherichia coli . Mol Cell. 2003;12:959–970. - PubMed
-
- Viswanathan A, You HJ, Doetsch PW. Phenotypic change caused by transcriptional bypass of uracil in nondividing cells. Science. 1999;284:159–162. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

