Nitrate-responsive miR393/AFB3 regulatory module controls root system architecture in Arabidopsis thaliana
- PMID: 20142497
- PMCID: PMC2840086
- DOI: 10.1073/pnas.0909571107
Nitrate-responsive miR393/AFB3 regulatory module controls root system architecture in Arabidopsis thaliana
Abstract
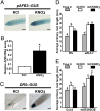
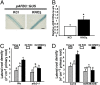
One of the most striking examples of plant developmental plasticity to changing environmental conditions is the modulation of root system architecture (RSA) in response to nitrate supply. Despite the fundamental and applied significance of understanding this process, the molecular mechanisms behind nitrate-regulated changes in developmental programs are still largely unknown. Small RNAs (sRNAs) have emerged as master regulators of gene expression in plants and other organisms. To evaluate the role of sRNAs in the nitrate response, we sequenced sRNAs from control and nitrate-treated Arabidopsis seedlings using the 454 sequencing technology. miR393 was induced by nitrate in these experiments. miR393 targets transcripts that code for a basic helix-loop-helix (bHLH) transcription factor and for the auxin receptors TIR1, AFB1, AFB2, and AFB3. However, only AFB3 was regulated by nitrate in roots under our experimental conditions. Analysis of the expression of this miR393/AFB3 module, revealed an incoherent feed-forward mechanism that is induced by nitrate and repressed by N metabolites generated by nitrate reduction and assimilation. To understand the functional role of this N-regulatory module for plant development, we analyzed the RSA response to nitrate in AFB3 insertional mutant plants and in miR393 overexpressors. RSA analysis in these plants revealed that both primary and lateral root growth responses to nitrate were altered. Interestingly, regulation of RSA by nitrate was specifically mediated by AFB3, indicating that miR393/AFB3 is a unique N-responsive module that controls root system architecture in response to external and internal N availability in Arabidopsis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

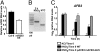
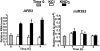
Similar articles
-
Systems approaches map regulatory networks downstream of the auxin receptor AFB3 in the nitrate response of Arabidopsis thaliana roots.Proc Natl Acad Sci U S A. 2013 Jul 30;110(31):12840-5. doi: 10.1073/pnas.1310937110. Epub 2013 Jul 11. Proc Natl Acad Sci U S A. 2013. PMID: 23847199 Free PMC article.
-
Regulation of auxin response by miR393-targeted transport inhibitor response protein 1 is involved in normal development in Arabidopsis.Plant Mol Biol. 2011 Dec;77(6):619-29. doi: 10.1007/s11103-011-9838-1. Epub 2011 Nov 1. Plant Mol Biol. 2011. PMID: 22042293
-
miR393 contributes to the embryogenic transition induced in vitro in Arabidopsis via the modification of the tissue sensitivity to auxin treatment.Planta. 2016 Jul;244(1):231-43. doi: 10.1007/s00425-016-2505-7. Epub 2016 Apr 4. Planta. 2016. PMID: 27040841 Free PMC article.
-
Signalling Overlaps between Nitrate and Auxin in Regulation of The Root System Architecture: Insights from the Arabidopsis thaliana.Int J Mol Sci. 2020 Apr 20;21(8):2880. doi: 10.3390/ijms21082880. Int J Mol Sci. 2020. PMID: 32326090 Free PMC article. Review.
-
Nitrate Signaling, Functions, and Regulation of Root System Architecture: Insights from Arabidopsis thaliana.Genes (Basel). 2020 Jun 9;11(6):633. doi: 10.3390/genes11060633. Genes (Basel). 2020. PMID: 32526869 Free PMC article. Review.
Cited by
-
Nitrogen and Stem Development: A Puzzle Still to Be Solved.Front Plant Sci. 2021 Feb 15;12:630587. doi: 10.3389/fpls.2021.630587. eCollection 2021. Front Plant Sci. 2021. PMID: 33659017 Free PMC article. Review.
-
Systems approaches map regulatory networks downstream of the auxin receptor AFB3 in the nitrate response of Arabidopsis thaliana roots.Proc Natl Acad Sci U S A. 2013 Jul 30;110(31):12840-5. doi: 10.1073/pnas.1310937110. Epub 2013 Jul 11. Proc Natl Acad Sci U S A. 2013. PMID: 23847199 Free PMC article.
-
Ectopic expression of miR160 results in auxin hypersensitivity, cytokinin hyposensitivity, and inhibition of symbiotic nodule development in soybean.Plant Physiol. 2013 Aug;162(4):2042-55. doi: 10.1104/pp.113.220699. Epub 2013 Jun 24. Plant Physiol. 2013. PMID: 23796794 Free PMC article.
-
Lateral root formation and nutrients: nitrogen in the spotlight.Plant Physiol. 2021 Nov 3;187(3):1104-1116. doi: 10.1093/plphys/kiab145. Plant Physiol. 2021. PMID: 33768243 Free PMC article. Review.
-
miRNomes involved in imparting thermotolerance to crop plants.3 Biotech. 2018 Dec;8(12):497. doi: 10.1007/s13205-018-1521-7. Epub 2018 Nov 24. 3 Biotech. 2018. PMID: 30498670 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

