Histidine-mediated RNA transfer to GDP for unique mRNA capping by vesicular stomatitis virus RNA polymerase
- PMID: 20142503
- PMCID: PMC2840475
- DOI: 10.1073/pnas.0913083107
Histidine-mediated RNA transfer to GDP for unique mRNA capping by vesicular stomatitis virus RNA polymerase
Abstract
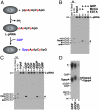
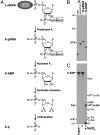
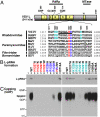
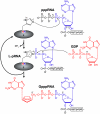
The RNA-dependent RNA polymerase L protein of vesicular stomatitis virus, a prototype of nonsegmented negative-strand (NNS) RNA viruses, forms a covalent complex with a 5'-phosphorylated viral mRNA-start sequence (L-pRNA), a putative intermediate in the unconventional mRNA capping reaction catalyzed by the RNA:GDP polyribonucleotidyltransferase (PRNTase) activity. Here, we directly demonstrate that the purified L-pRNA complex transfers pRNA to GDP to produce the capped RNA (Gpp-pRNA), indicating that the complex is a bona fide intermediate in the RNA transfer reaction. To locate the active site of the PRNTase domain in the L protein, the covalent RNA attachment site was mapped. We found that the 5'-monophosphate end of the RNA is linked to the histidine residue at position 1,227 (H1227) of the L protein through a phosphoamide bond. Interestingly, H1227 is part of the histidine-arginine (HR) motif, which is conserved within the L proteins of the NNS RNA viruses including rabies, measles, Ebola, and Borna disease viruses. Mutagenesis analyses revealed that the HR motif is required for the PRNTase activity at the step of the enzyme-pRNA intermediate formation. Thus, our findings suggest that an ancient NNS RNA viral polymerase has acquired the PRNTase domain independently of the eukaryotic mRNA capping enzyme during evolution and PRNTase becomes a rational target for designing antiviral agents.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Comment in
-
Viruses know more than one way to don a cap.Proc Natl Acad Sci U S A. 2010 Feb 23;107(8):3283-4. doi: 10.1073/pnas.0915061107. Epub 2010 Feb 18. Proc Natl Acad Sci U S A. 2010. PMID: 20167804 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

