Mechanism of chromatin remodeling
- PMID: 20142505
- PMCID: PMC2817641
- DOI: 10.1073/pnas.1000398107
Mechanism of chromatin remodeling
Abstract
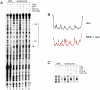
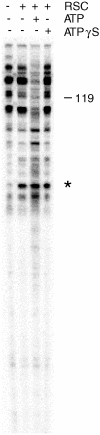
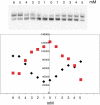
Results from biochemical and structural studies of the RSC chromatin-remodeling complex prompt a proposal for the remodeling mechanism: RSC binding to the nucleosome releases the DNA from the histone surface and initiates DNA translocation (through one or a small number of DNA base pairs); ATP binding completes translocation, and ATP hydrolysis resets the system. Binding energy thus plays a central role in the remodeling process. RSC may disrupt histone-DNA contacts by affecting histone octamer conformation and through extensive interaction with the DNA. Bulging of the DNA from the octamer surface is possible, and twisting is unavoidable, but neither is the basis of remodeling.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Lorch Y, Zhang M, Kornberg RD. Histone octamer transfer by a chromatin-remodeling complex. Cell. 1999;96(3):389–392. - PubMed
-
- Whitehouse I, et al. Nucleosome mobilization catalysed by the yeast SWI/SNF complex. Nature. 1999;400(6746):784–787. - PubMed
-
- Saha A, Wittmeyer J, Cairns BR. Chromatin remodelling: The industrial revolution of DNA around histones. Nat Rev Mol Cell Biol. 2006;7(6):437–447. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

