A statistically rigorous test for the identification of parent-fragment pairs in LC-MS datasets
- PMID: 20143830
- PMCID: PMC2829950
- DOI: 10.1021/ac902361f
A statistically rigorous test for the identification of parent-fragment pairs in LC-MS datasets
Abstract
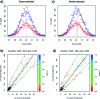
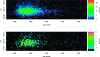
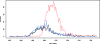
Untargeted global metabolic profiling by liquid chromatography-mass spectrometry generates numerous signals that are due to unknown compounds and whose identification forms an important challenge. The analysis of metabolite fragmentation patterns, following collision-induced dissociation, provides a valuable tool for identification, but can be severely impeded by close chromatographic coelution of distinct metabolites. We propose a new algorithm for identifying related parent-fragment pairs and for distinguishing these from signals due to unrelated compounds. Unlike existing methods, our approach addresses the problem by means of a hypothesis test that is based on the distribution of the recorded ion counts, and thereby provides a statistically rigorous measure of the uncertainty involved in the classification problem. Because of technological constraints, the test is of primary use at low and intermediate ion counts, above which detector saturation causes substantial bias to the recorded ion count. The validity of the test is demonstrated through its application to pairs of coeluting isotopologues and to known parent-fragment pairs, which results in test statistics consistent with the null distribution. The performance of the test is compared with a commonly used Pearson correlation approach and found to be considerably better (e.g., false positive rate of 6.25%, compared with a value of 50% for the correlation for perfectly coeluting ions). Because the algorithm may be used for the analysis of high-mass compounds in addition to metabolic data, we expect it to facilitate the analysis of fragmentation patterns for a wide range of analytical problems.
Figures






References
-
- Raamsdonk L. M.; Teusink B.; Broadhurst D.; Zhang N. S.; Hayes A.; Walsh M. C.; Berden J. A.; Brindle K. M.; Kell D. B.; Rowland J. J.; Westerhoff H. V.; van Dam K.; Oliver S. G. Nat. Biotechnol. 2001, 19, 45–50. - PubMed
-
- Nicholson J. K.; Lindon J. C.; Holmes E. Xenobiotica 1999, 29 (11), 1181–1189. - PubMed
-
- Want E. J.; Nordstrom A.; Morita H.; Siuzdak G. J. Proteome Res. 2007, 6, 459–468. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

