Genomic and evolutionary comparisons of diazotrophic and pathogenic bacteria of the order Rhizobiales
- PMID: 20144182
- PMCID: PMC2907836
- DOI: 10.1186/1471-2180-10-37
Genomic and evolutionary comparisons of diazotrophic and pathogenic bacteria of the order Rhizobiales
Abstract
Background: Species belonging to the Rhizobiales are intriguing and extensively researched for including both bacteria with the ability to fix nitrogen when in symbiosis with leguminous plants and pathogenic bacteria to animals and plants. Similarities between the strategies adopted by pathogenic and symbiotic Rhizobiales have been described, as well as high variability related to events of horizontal gene transfer. Although it is well known that chromosomal rearrangements, mutations and horizontal gene transfer influence the dynamics of bacterial genomes, in Rhizobiales, the scenario that determine pathogenic or symbiotic lifestyle are not clear and there are very few studies of comparative genomic between these classes of prokaryotic microorganisms trying to delineate the evolutionary characterization of symbiosis and pathogenesis.
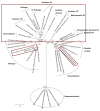
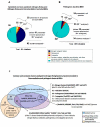
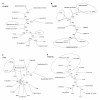
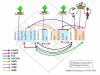
Results: Non-symbiotic nitrogen-fixing bacteria and bacteria involved in bioremediation closer to symbionts and pathogens in study may assist in the origin and ancestry genes and the gene flow occurring in Rhizobiales. The genomic comparisons of 19 species of Rhizobiales, including nitrogen-fixing, bioremediators and pathogens resulted in 33 common clusters to biological nitrogen fixation and pathogenesis, 15 clusters exclusive to all nitrogen-fixing bacteria and bacteria involved in bioremediation, 13 clusters found in only some nitrogen-fixing and bioremediation bacteria, 01 cluster exclusive to some symbionts, and 01 cluster found only in some pathogens analyzed. In BBH performed to all strains studied, 77 common genes were obtained, 17 of which were related to biological nitrogen fixation and pathogenesis. Phylogenetic reconstructions for Fix, Nif, Nod, Vir, and Trb showed possible horizontal gene transfer events, grouping species of different phenotypes.
Conclusions: The presence of symbiotic and virulence genes in both pathogens and symbionts does not seem to be the only determinant factor for lifestyle evolution in these microorganisms, although they may act in common stages of host infection. The phylogenetic analysis for many distinct operons involved in these processes emphasizes the relevance of horizontal gene transfer events in the symbiotic and pathogenic similarity.
Figures
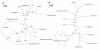
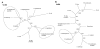



References
-
- Kaneko T, Nakamura Y, Sato S, Asamizu E, Kato T, Sasamoto S, Watanabe A, Idesawa K, Ishikawa A, Kawashima K, Kimura T, Kishida Y, Kiyokawa C, Kohara M, Matsumoto M, Matsuno A, Mochizuki Y, Nakayama S, Nakazaki N, Shimpo S, Sugimoto M, Takeuchi C, Yamada M, Tabata S. Complete genome structure of the nitrogen-fixing symbiotic bacterium Mesorhizobium loti. DNA Res. 2000;7:331–338. doi: 10.1093/dnares/7.6.331. - DOI - PubMed
-
- Paulsen IT, Seshadri R, Nelson KE, Eisen JA, Heidelberg JF, Read TD, Dodson RJ, Umayam L, Brinkac LM, Beanan MJ, Daugherty SC, Deboy RT, Durkin AS, Kolonay JF, Madupu R, Nelson WC, Ayodeji B, Kraul M, Shetty J, Malek J, Van Aken SE, Riedmuller S, Tettelin H, Gill SR, White O, Salzberg SL, Hoover DL, Lindler LE, Halling SM, Boyle SM, Fraser CM. The Brucella suis genome reveals fundamental similarities between animal and plant pathogens and symbionts. Proc Natl Acad Sci USA. 2002;99:13148–13153. doi: 10.1073/pnas.192319099. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

