Annotation and classification of the bovine T cell receptor delta genes
- PMID: 20144200
- PMCID: PMC2846910
- DOI: 10.1186/1471-2164-11-100
Annotation and classification of the bovine T cell receptor delta genes
Abstract
Background: gammadelta T cells differ from alphabeta T cells with regard to the types of antigen with which their T cell receptors interact; gammadelta T cell antigens are not necessarily peptides nor are they presented on MHC. Cattle are considered a "gammadelta T cell high" species indicating they have an increased proportion of gammadelta T cells in circulation relative to that in "gammadelta T cell low" species such as humans and mice. Prior to the onset of the studies described here, there was limited information regarding the genes that code for the T cell receptor delta chains of this gammadelta T cell high species.
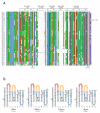
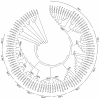
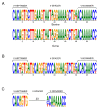
Results: By annotating the bovine (Bos taurus) genome Btau_3.1 assembly the presence of 56 distinct T cell receptor delta (TRD) variable (V) genes were found, 52 of which belong to the TRDV1 subgroup and were co-mingled with the T cell receptor alpha variable (TRAV) genes. In addition, two genes belonging to the TRDV2 subgroup and single TRDV3 and TRDV4 genes were found. We confirmed the presence of five diversity (D) genes, three junctional (J) genes and a single constant (C) gene and describe the organization of the TRD locus. The TRDV4 gene is found downstream of the C gene and in an inverted orientation of transcription, consistent with its orthologs in humans and mice. cDNA evidence was assessed to validate expression of the variable genes and showed that one to five D genes could be incorporated into a single transcript. Finally, we grouped the bovine and ovine TRDV1 genes into sets based on their relatedness.
Conclusions: The bovine genome contains a large and diverse repertoire of TRD genes when compared to the genomes of "gammadelta T cell low" species. This suggests that in cattle gammadelta T cells play a more important role in immune function since they would be predicted to bind a greater variety of antigens.
Figures




Similar articles
-
Sheep (Ovis aries) T cell receptor alpha (TRA) and delta (TRD) genes and genomic organization of the TRA/TRD locus.BMC Genomics. 2015 Sep 18;16:709. doi: 10.1186/s12864-015-1790-z. BMC Genomics. 2015. PMID: 26383271 Free PMC article.
-
Identification of three new bovine T-cell receptor delta variable gene subgroups expressed by peripheral blood T cells.Immunogenetics. 2006 Sep;58(9):746-57. doi: 10.1007/s00251-006-0136-z. Epub 2006 Aug 1. Immunogenetics. 2006. PMID: 16896832
-
Genomic analysis offers insights into the evolution of the bovine TRA/TRD locus.BMC Genomics. 2014 Nov 19;15(1):994. doi: 10.1186/1471-2164-15-994. BMC Genomics. 2014. PMID: 25408163 Free PMC article.
-
Bovine gamma delta T cells and the function of gamma delta T cell specific WC1 co-receptors.Cell Immunol. 2015 Jul;296(1):76-86. doi: 10.1016/j.cellimm.2015.05.003. Epub 2015 May 16. Cell Immunol. 2015. PMID: 26008759 Review.
-
The bovine model for elucidating the role of γδ T cells in controlling infectious diseases of importance to cattle and humans.Mol Immunol. 2015 Jul;66(1):35-47. doi: 10.1016/j.molimm.2014.10.024. Epub 2014 Dec 26. Mol Immunol. 2015. PMID: 25547715 Review.
Cited by
-
Spectratype analysis of the T cell receptor δ CDR3 region of bovine γδ T cells responding to leptospira.Immunogenetics. 2015 Feb;67(2):95-109. doi: 10.1007/s00251-014-0817-y. Epub 2014 Dec 12. Immunogenetics. 2015. PMID: 25502871
-
Genomic Variants Revealed by Invariably Missing Genotypes in Nelore Cattle.PLoS One. 2015 Aug 25;10(8):e0136035. doi: 10.1371/journal.pone.0136035. eCollection 2015. PLoS One. 2015. PMID: 26305794 Free PMC article.
-
A comprehensive analysis of the germline and expressed TCR repertoire in White Peking duck.Sci Rep. 2017 Jan 30;7:41426. doi: 10.1038/srep41426. Sci Rep. 2017. PMID: 28134319 Free PMC article.
-
IMGT®Homo sapiens IG and TR Loci, Gene Order, CNV and Haplotypes: New Concepts as a Paradigm for Jawed Vertebrates Genome Assemblies.Biomolecules. 2022 Feb 28;12(3):381. doi: 10.3390/biom12030381. Biomolecules. 2022. PMID: 35327572 Free PMC article.
-
Characterization of the domestic goat γδ T cell receptor gene loci and gene usage.Immunogenetics. 2021 Apr;73(2):187-201. doi: 10.1007/s00251-021-01203-y. Epub 2021 Jan 21. Immunogenetics. 2021. PMID: 33479855
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

