Stone formation in peach fruit exhibits spatial coordination of the lignin and flavonoid pathways and similarity to Arabidopsis dehiscence
- PMID: 20144217
- PMCID: PMC2830173
- DOI: 10.1186/1741-7007-8-13
Stone formation in peach fruit exhibits spatial coordination of the lignin and flavonoid pathways and similarity to Arabidopsis dehiscence
Abstract
Background: Lignification of the fruit endocarp layer occurs in many angiosperms and plays a critical role in seed protection and dispersal. This process has been extensively studied with relationship to pod shatter or dehiscence in Arabidopsis. Dehiscence is controlled by a set of transcription factors that define the fruit tissue layers and whether or not they lignify. In contrast, relatively little is known about similar processes in other plants such as stone fruits which contain an extremely hard lignified endocarp or stone surrounding a single seed.
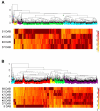
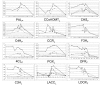
Results: Here we show that lignin deposition in peach initiates near the blossom end within the endocarp layer and proceeds in a distinct spatial-temporal pattern. Microarray studies using a developmental series from young fruits identified a sharp and transient induction of phenylpropanoid, lignin and flavonoid pathway genes concurrent with lignification and subsequent stone hardening. Quantitative polymerase chain reaction studies revealed that specific phenylpropanoid (phenylalanine ammonia-lyase and cinnamate 4-hydroxylase) and lignin (caffeoyl-CoA O-methyltransferase, peroxidase and laccase) pathway genes were induced in the endocarp layer over a 10 day time period, while two lignin genes (p-coumarate 3-hydroxylase and cinnamoyl CoA reductase) were co-regulated with flavonoid pathway genes (chalcone synthase, dihydroflavanol 4-reductase, leucoanthocyanidin dioxygen-ase and flavanone-3-hydrosylase) which were mesocarp and exocarp specific. Analysis of other fruit development expression studies revealed that flavonoid pathway induction is conserved in the related Rosaceae species apple while lignin pathway induction is not. The transcription factor expression of peach genes homologous to known endocarp determinant genes in Arabidopsis including SHATTERPROOF, SEEDSTCK and NAC SECONDARY WALL THICENING PROMOTING FACTOR 1 were found to be specifically expressed in the endocarp while the negative regulator FRUITFUL predominated in exocarp and mesocarp.
Conclusions: Collectively, the data suggests, first, that the process of endocarp determination and differentiation in peach and Arabidopsis share common regulators and, secondly, reveals a previously unknown coordination of competing lignin and flavonoid biosynthetic pathways during early fruit development.
Figures




References
-
- Ryugo K. The rate of dry weight accumulation by the peach pit during the hardening process. Amer Soc Hort Sci. 1961;78:132–137.
-
- Ryugo K. Changes in methoxyl content in the peach endocarp and some of its soluble phenolic constituents during lignification. Amer Soc Hort Sci. 1963;84:110–115.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

