Dynamic probe selection for studying microbial transcriptome with high-density genomic tiling microarrays
- PMID: 20144223
- PMCID: PMC2836303
- DOI: 10.1186/1471-2105-11-82
Dynamic probe selection for studying microbial transcriptome with high-density genomic tiling microarrays
Abstract
Background: Current commercial high-density oligonucleotide microarrays can hold millions of probe spots on a single microscopic glass slide and are ideal for studying the transcriptome of microbial genomes using a tiling probe design. This paper describes a comprehensive computational pipeline implemented specifically for designing tiling probe sets to study microbial transcriptome profiles.
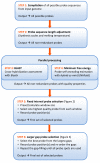
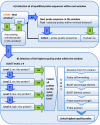
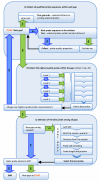
Results: The pipeline identifies every possible probe sequence from both forward and reverse-complement strands of all DNA sequences in the target genome including circular or linear chromosomes and plasmids. Final probe sequence lengths are adjusted based on the maximal oligonucleotide synthesis cycles and best isothermality allowed. Optimal probes are then selected in two stages - sequential and gap-filling. In the sequential stage, probes are selected from sequence windows tiled alongside the genome. In the gap-filling stage, additional probes are selected from the largest gaps between adjacent probes that have already been selected, until a predefined number of probes is reached. Selection of the highest quality probe within each window and gap is based on five criteria: sequence uniqueness, probe self-annealing, melting temperature, oligonucleotide length, and probe position.
Conclusions: The probe selection pipeline evaluates global and local probe sequence properties and selects a set of probes dynamically and evenly distributed along the target genome. Unique to other similar methods, an exact number of non-redundant probes can be designed to utilize all the available probe spots on any chosen microarray platform. The pipeline can be applied to microbial genomes when designing high-density tiling arrays for comparative genomics, ChIP chip, gene expression and comprehensive transcriptome studies.
Figures
Similar articles
-
Thermodynamically optimal whole-genome tiling microarray design and validation.BMC Res Notes. 2016 Jun 13;9:305. doi: 10.1186/s13104-016-2113-4. BMC Res Notes. 2016. PMID: 27295952 Free PMC article.
-
Efficient oligonucleotide probe selection for pan-genomic tiling arrays.BMC Bioinformatics. 2009 Sep 16;10:293. doi: 10.1186/1471-2105-10-293. BMC Bioinformatics. 2009. PMID: 19758451 Free PMC article.
-
UPS 2.0: unique probe selector for probe design and oligonucleotide microarrays at the pangenomic/genomic level.BMC Genomics. 2010 Dec 2;11 Suppl 4(Suppl 4):S6. doi: 10.1186/1471-2164-11-S4-S6. BMC Genomics. 2010. PMID: 21143815 Free PMC article.
-
In-depth query of large genomes using tiling arrays.Methods Mol Biol. 2007;377:163-74. doi: 10.1007/978-1-59745-390-5_10. Methods Mol Biol. 2007. PMID: 17634616 Review.
-
Applications of DNA tiling arrays for whole-genome analysis.Genomics. 2005 Jan;85(1):1-15. doi: 10.1016/j.ygeno.2004.10.005. Genomics. 2005. PMID: 15607417 Review.
Cited by
-
Roseomics: a blank slate.Curr Opin Virol. 2014 Dec;9:188-93. doi: 10.1016/j.coviro.2014.09.021. Epub 2014 Nov 28. Curr Opin Virol. 2014. PMID: 25437230 Free PMC article. Review.
-
Microarray oligonucleotide probe designer (MOPeD): A web service.Open Access Bioinformatics. 2010 Nov 1;2(2010):145-155. doi: 10.2147/OAB.S13741. Open Access Bioinformatics. 2010. PMID: 21379402 Free PMC article.
-
Thermodynamically optimal whole-genome tiling microarray design and validation.BMC Res Notes. 2016 Jun 13;9:305. doi: 10.1186/s13104-016-2113-4. BMC Res Notes. 2016. PMID: 27295952 Free PMC article.
-
Comprehensive transcriptome analysis of the periodontopathogenic bacterium Porphyromonas gingivalis W83.J Bacteriol. 2012 Jan;194(1):100-14. doi: 10.1128/JB.06385-11. Epub 2011 Oct 28. J Bacteriol. 2012. PMID: 22037400 Free PMC article.
-
A hidden Markov support vector machine framework incorporating profile geometry learning for identifying microbial RNA in tiling array data.Bioinformatics. 2010 Jun 1;26(11):1423-30. doi: 10.1093/bioinformatics/btq162. Epub 2010 Apr 15. Bioinformatics. 2010. PMID: 20395286 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

