Molecular risk assessment of BIG 1-98 participants by expression profiling using RNA from archival tissue
- PMID: 20144231
- PMCID: PMC2829498
- DOI: 10.1186/1471-2407-10-37
Molecular risk assessment of BIG 1-98 participants by expression profiling using RNA from archival tissue
Abstract
Background: The purpose of the work reported here is to test reliable molecular profiles using routinely processed formalin-fixed paraffin-embedded (FFPE) tissues from participants of the clinical trial BIG 1-98 with a median follow-up of 60 months.
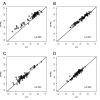
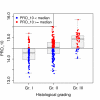
Methods: RNA from fresh frozen (FF) and FFPE tumor samples of 82 patients were used for quality control, and independent FFPE tissues of 342 postmenopausal participants of BIG 1-98 with ER-positive cancer were analyzed by measuring prospectively selected genes and computing scores representing the functions of the estrogen receptor (eight genes, ER_8), the progesterone receptor (five genes, PGR_5), Her2 (two genes, HER2_2), and proliferation (ten genes, PRO_10) by quantitative reverse transcription PCR (qRT-PCR) on TaqMan Low Density Arrays. Molecular scores were computed for each category and ER_8, PGR_5, HER2_2, and PRO_10 scores were combined into a RISK_25 score.
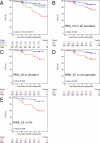
Results: Pearson correlation coefficients between FF- and FFPE-derived scores were at least 0.94 and high concordance was observed between molecular scores and immunohistochemical data. The HER2_2, PGR_5, PRO_10 and RISK_25 scores were significant predictors of disease free-survival (DFS) in univariate Cox proportional hazard regression. PRO_10 and RISK_25 scores predicted DFS in patients with histological grade II breast cancer and in lymph node positive disease. The PRO_10 and PGR_5 scores were independent predictors of DFS in multivariate Cox regression models incorporating clinical risk indicators; PRO_10 outperformed Ki-67 labeling index in multivariate Cox proportional hazard analyses.
Conclusions: Scores representing the endocrine responsiveness and proliferation status of breast cancers were developed from gene expression analyses based on RNA derived from FFPE tissues. The validation of the molecular scores with tumor samples of participants of the BIG 1-98 trial demonstrates that such scores can serve as independent prognostic factors to estimate disease free survival (DFS) in postmenopausal patients with estrogen receptor positive breast cancer.
Trial registration: ClinicalTrials.gov NCT00004205.
Figures

Similar articles
-
Clinical relevance of Ki67 gene expression analysis using formalin-fixed paraffin-embedded breast cancer specimens.Breast Cancer. 2013 Jul;20(3):262-70. doi: 10.1007/s12282-012-0332-7. Epub 2012 Feb 24. Breast Cancer. 2013. PMID: 22362219
-
Investigation of recurrence prediction ability of EndoPredict® using microarray data from fresh frozen tissues in ER-positive, HER2-negative breast cancer and indication expansion of EndoPredict® from microarray data from fresh-frozen to FFPE tissues.Breast Cancer. 2024 Jul;31(4):593-606. doi: 10.1007/s12282-024-01573-7. Epub 2024 Apr 8. Breast Cancer. 2024. PMID: 38587783
-
72-gene classifier for predicting prognosis of estrogen receptor-positive and node-negative breast cancer patients using formalin-fixed, paraffin-embedded tumor tissues.Clin Breast Cancer. 2014 Jun;14(3):e73-80. doi: 10.1016/j.clbc.2013.11.006. Epub 2013 Nov 22. Clin Breast Cancer. 2014. PMID: 24461457
-
Gene expression of estrogen receptor, progesterone receptor and microtubule-associated protein Tau in high-risk early breast cancer: a quest for molecular predictors of treatment benefit in the context of a Hellenic Cooperative Oncology Group trial.Breast Cancer Res Treat. 2009 Jul;116(1):131-43. doi: 10.1007/s10549-008-0144-9. Epub 2008 Jul 31. Breast Cancer Res Treat. 2009. PMID: 18668363 Clinical Trial.
-
The Visualization of Protein-Protein Interactions in Breast Cancer: Deployment Study in Pathological Examination.Acta Histochem Cytochem. 2021 Dec 24;54(6):177-183. doi: 10.1267/ahc.21-00084. Epub 2021 Nov 18. Acta Histochem Cytochem. 2021. PMID: 35023880 Free PMC article. Review.
Cited by
-
Joint analysis of histopathology image features and gene expression in breast cancer.BMC Bioinformatics. 2016 May 11;17(1):209. doi: 10.1186/s12859-016-1072-z. BMC Bioinformatics. 2016. PMID: 27170365 Free PMC article.
-
Reliable PCR quantitation of estrogen, progesterone and ERBB2 receptor mRNA from formalin-fixed, paraffin-embedded tissue is independent of prior macro-dissection.Virchows Arch. 2013 Dec;463(6):775-86. doi: 10.1007/s00428-013-1486-1. Epub 2013 Oct 8. Virchows Arch. 2013. PMID: 24100522
-
Cancer cell-autonomous contribution of type I interferon signaling to the efficacy of chemotherapy.Nat Med. 2014 Nov;20(11):1301-9. doi: 10.1038/nm.3708. Epub 2014 Oct 26. Nat Med. 2014. PMID: 25344738 Clinical Trial.
-
Characterization of molecular scores and gene expression signatures in primary breast cancer, local recurrences and brain metastases.BMC Cancer. 2019 Jun 7;19(1):549. doi: 10.1186/s12885-019-5752-8. BMC Cancer. 2019. PMID: 31174485 Free PMC article.
References
-
- Carlson RW, Jahanzeb M, Kiel K, Marks LB, Mc Cromick B, Pierce LJ, Ward JH, Topham NS. NCCN Clinical Practice Guidelines in Oncology V.2.2008. Book NCCN Clinical Practice Guidelines in Oncology V.2. 2008. http://www.nccn.org
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

