Genome-wide methylation and expression profiling identifies promoter characteristics affecting demethylation-induced gene up-regulation in melanoma
- PMID: 20144234
- PMCID: PMC2843643
- DOI: 10.1186/1755-8794-3-4
Genome-wide methylation and expression profiling identifies promoter characteristics affecting demethylation-induced gene up-regulation in melanoma
Abstract
Background: Abberant DNA methylation at CpG dinucleotides represents a common mechanism of transcriptional silencing in cancer. Since CpG methylation is a reversible event, tumor supressor genes that have undergone silencing through this mechanism represent promising targets for epigenetically active anti-cancer therapy. The cytosine analog 5-aza-2'-deoxycytidine (decitabine) induces genomic hypomethylation by inhibiting DNA methyltransferase, and is an example of an epigenetic agent that is thought to act by up-regulating silenced genes.
Methods: It is unclear why decitabine causes some silenced loci to re-express, while others remain inactive. By applying data-mining techniques to large-scale datasets, we attempted to elucidate the qualities of promoter regions that define susceptibility to the drug's action. Our experimental data, derived from melanoma cell strains, consist of genome-wide gene expression data before and after treatment with decitabine, as well as genome-wide data on un-treated promoter methylation status, and validation of specific genes by bisulfite sequencing.
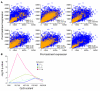
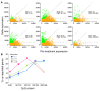
Results: We show that the combination of promoter CpG content and methylation level informs the ability of decitabine treatment to up-regulate gene expression. Promoters with high methylation levels and intermediate CpG content appear most susceptible to up-regulation by decitabine, whereas few of those highly methylated promoters with high CpG content are up-regulated. For promoters with low methylation levels, those with high CpG content are more likely to be up-regulated, whereas those with low CpG content are underrepresented among up-regulated genes.
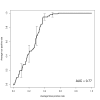
Conclusions: Clinically, elucidating the patterns of action of decitabine could aid in predicting the likelihood of up-regulating epigenetically silenced tumor suppressor genes and others from pathways involved with tumor biology. As a first step toward an eventual translational application, we build a classifier to predict gene up-regulation based on promoter methylation and CpG content, which achieves a performance of 0.77 AUC.
Figures

Similar articles
-
Demethylation by 5-aza-2'-deoxycytidine in colorectal cancer cells targets genomic DNA whilst promoter CpG island methylation persists.BMC Cancer. 2010 Jul 12;10:366. doi: 10.1186/1471-2407-10-366. BMC Cancer. 2010. PMID: 20618997 Free PMC article.
-
NGX6 gene mediated by promoter methylation as a potential molecular marker in colorectal cancer.BMC Cancer. 2010 Apr 27;10:160. doi: 10.1186/1471-2407-10-160. BMC Cancer. 2010. PMID: 20423473 Free PMC article.
-
Gemcitabine reactivates epigenetically silenced genes and functions as a DNA methyltransferase inhibitor.Int J Mol Med. 2012 Dec;30(6):1505-11. doi: 10.3892/ijmm.2012.1138. Epub 2012 Sep 21. Int J Mol Med. 2012. PMID: 23007409
-
Modes of action of the DNA methyltransferase inhibitors azacytidine and decitabine.Int J Cancer. 2008 Jul 1;123(1):8-13. doi: 10.1002/ijc.23607. Int J Cancer. 2008. PMID: 18425818 Review.
-
Epigenetic DNA-(cytosine-5-carbon) modifications: 5-aza-2'-deoxycytidine and DNA-demethylation.Biochemistry (Mosc). 2009 Jun;74(6):613-9. doi: 10.1134/s0006297909060042. Biochemistry (Mosc). 2009. PMID: 19645665 Review.
Cited by
-
Integrating Early Transcriptomic Responses to Rhizotoxins in Rice (Oryza sativa. L.) Reveals Key Regulators and a Potential Early Biomarker of Cadmium Toxicity.Front Plant Sci. 2017 Aug 18;8:1432. doi: 10.3389/fpls.2017.01432. eCollection 2017. Front Plant Sci. 2017. PMID: 28868059 Free PMC article.
-
Expression of proteins involved in epigenetic regulation in human cutaneous melanoma and peritumoral skin.Tumour Biol. 2014 Aug;35(8):8225-33. doi: 10.1007/s13277-014-2098-3. Epub 2014 May 22. Tumour Biol. 2014. PMID: 24850177
-
A semantic web framework to integrate cancer omics data with biological knowledge.BMC Bioinformatics. 2012 Jan 25;13 Suppl 1(Suppl 1):S10. doi: 10.1186/1471-2105-13-S1-S10. BMC Bioinformatics. 2012. PMID: 22373303 Free PMC article.
-
Augmenting chemotherapy with low-dose decitabine through an immune-independent mechanism.JCI Insight. 2022 Nov 22;7(22):e159419. doi: 10.1172/jci.insight.159419. JCI Insight. 2022. PMID: 36227698 Free PMC article.
-
Genome-scale DNA methylation pattern profiling of human bone marrow mesenchymal stem cells in long-term culture.Exp Mol Med. 2012 Aug 31;44(8):503-12. doi: 10.3858/emm.2012.44.8.057. Exp Mol Med. 2012. PMID: 22684242 Free PMC article.
References
-
- Gonzalez-Zulueta M, Bender C, Yang A, Nguyen T, Beart R, Van Tornout J, Jones P. Methylation of the 5' CpG island of the p16/CDKN2 tumor suppressor gene in normal and transformed human tissues correlates with gene silencing. Cancer Res. 1995;55(20):4531–4535. - PubMed
-
- Martinez R, Martin-Subero J, Rohde V, Kirsch M, Alaminos M, Fernandez A, Ropero S, Schackert G, Esteller M. A microarray-based DNA methylation study of glioblastoma multiforme. Epigenetics. 2009;4(4):255–264. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

