Cyclooxygenase-2 is a novel transcriptional target of the nuclear EGFR-STAT3 and EGFRvIII-STAT3 signaling axes
- PMID: 20145033
- PMCID: PMC2824777
- DOI: 10.1158/1541-7786.MCR-09-0391
Cyclooxygenase-2 is a novel transcriptional target of the nuclear EGFR-STAT3 and EGFRvIII-STAT3 signaling axes
Abstract
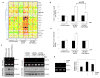
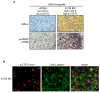
Emerging evidence indicates a novel mode of epidermal growth factor receptor (EGFR) signaling, notably, one involves EGFR nuclear translocalization and subsequent gene activation. To date, however, the significance of the nuclear EGFR pathway in glioblastoma (GBM) is unknown. Here, we report that EGFR and its constitutively activated variant EGFRvIII undergo nuclear translocalization in GBM cells, in which the former event requires EGF stimulation and the latter is constitutive. To gain insights into the effect of nuclear EGFR on gene expression in GBM, we created isogenic GBM cell lines, namely, U87MG-vector, U87MG-EGFR, and U87MG-EGFRdNLS that, respectively, express the control vector, EGFR, and nuclear entry-defective EGFR with a deletion of the nuclear localization signal (NLS). Microarray analysis shows that 19 genes, including cyclooxygenase-2 (COX-2), to be activated in U87MG-EGFR cells but not in U87MG-EGFRdNLS and U87MG-vector cells. Subsequent validation studies indicate that COX-2 gene is expressed at higher levels in cells with EGFR and EGFRvIII than those with EGFRdNLS and EGFRvIIIdNLS. Nuclear EGFR and its transcriptional cofactor signal transducer and activator of transcription 3 (STAT3) associate with the COX-2 promoter. Increased expression of EGFR/EGFRvIII and activated STAT3 leads to the synergistic activation of the COX-2 promoter. Promoter mutational analysis identified a proximal STAT3-binding site that is required for EGFR/EGFRvIII-STAT3-mediated COX-2 gene activation. In GBM tumors, an association exists between levels of COX-2, EGFR/EGFRvIII, and activated STAT3. Together, these findings indicate the existence of the nuclear EGFR/EGFRvIII signaling pathway in GBM and its functional interaction with STAT3 to activate COX-2 gene expression, thus linking EGFR-STAT3 and EGFRvIII-STAT3 signaling axes to proinflammatory COX-2 mediated pathway.
Figures




Similar articles
-
STAT1 gene expression is enhanced by nuclear EGFR and HER2 via cooperation with STAT3.Mol Carcinog. 2013 Dec;52(12):959-69. doi: 10.1002/mc.21936. Epub 2012 Jun 12. Mol Carcinog. 2013. PMID: 22693070 Free PMC article.
-
JAK2/STAT3 targeted therapy suppresses tumor invasion via disruption of the EGFRvIII/JAK2/STAT3 axis and associated focal adhesion in EGFRvIII-expressing glioblastoma.Neuro Oncol. 2014 Sep;16(9):1229-43. doi: 10.1093/neuonc/nou046. Epub 2014 May 25. Neuro Oncol. 2014. PMID: 24861878 Free PMC article.
-
EGFR phosphorylates tumor-derived EGFRvIII driving STAT3/5 and progression in glioblastoma.Cancer Cell. 2013 Oct 14;24(4):438-49. doi: 10.1016/j.ccr.2013.09.004. Cancer Cell. 2013. PMID: 24135280 Free PMC article.
-
EGFR-targeted therapy in malignant glioma: novel aspects and mechanisms of drug resistance.Curr Mol Pharmacol. 2010 Jan;3(1):37-52. doi: 10.2174/1874467211003010037. Curr Mol Pharmacol. 2010. PMID: 20030624 Free PMC article. Review.
-
Signal transducer and activator of transcription-3: a molecular hub for signaling pathways in gliomas.Mol Cancer Res. 2008 May;6(5):675-84. doi: 10.1158/1541-7786.MCR-07-2180. Mol Cancer Res. 2008. PMID: 18505913 Free PMC article. Review.
Cited by
-
Chronic Anatabine Treatment Reduces Alzheimer's Disease (AD)-Like Pathology and Improves Socio-Behavioral Deficits in a Transgenic Mouse Model of AD.PLoS One. 2015 May 26;10(5):e0128224. doi: 10.1371/journal.pone.0128224. eCollection 2015. PLoS One. 2015. PMID: 26010758 Free PMC article.
-
Synthesis and Significance of Arachidonic Acid, a Substrate for Cyclooxygenases, Lipoxygenases, and Cytochrome P450 Pathways in the Tumorigenesis of Glioblastoma Multiforme, Including a Pan-Cancer Comparative Analysis.Cancers (Basel). 2023 Feb 2;15(3):946. doi: 10.3390/cancers15030946. Cancers (Basel). 2023. PMID: 36765904 Free PMC article. Review.
-
Radiosensitivity enhancement by combined treatment of nimotuzumab and celecoxib on nasopharyngeal carcinoma cells.Drug Des Devel Ther. 2018 Jul 16;12:2223-2231. doi: 10.2147/DDDT.S163595. eCollection 2018. Drug Des Devel Ther. 2018. PMID: 30038488 Free PMC article.
-
RNA-binding protein p54nrb/NONO potentiates nuclear EGFR-mediated tumorigenesis of triple-negative breast cancer.Cell Death Dis. 2022 Jan 10;13(1):42. doi: 10.1038/s41419-021-04488-9. Cell Death Dis. 2022. PMID: 35013116 Free PMC article.
-
Non-canonical signaling mode of the epidermal growth factor receptor family.Am J Cancer Res. 2015 Sep 15;5(10):2944-58. eCollection 2015. Am J Cancer Res. 2015. PMID: 26693051 Free PMC article. Review.
References
-
- Nakamura JL. The epidermal growth factor receptor in malignant gliomas: pathogenesis and therapeutic implications. Expert Opinion on Therapeutic Targets. 2007;11:463. - PubMed
-
- Yang Z, Bagheri-Yarmand R, Wang RA, et al. The epidermal growth factor receptor tyrosine kinase inhibitor ZD1839 (Iressa) suppresses c-Src and Pak1 pathways and invasiveness of human cancer cells. Clin Cancer Res. 2004;10:658–67. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

