Urinary-type plasminogen activator receptor/alpha 3 beta 1 integrin signaling, altered gene expression, and oral tumor progression
- PMID: 20145038
- PMCID: PMC2825738
- DOI: 10.1158/1541-7786.MCR-09-0045
Urinary-type plasminogen activator receptor/alpha 3 beta 1 integrin signaling, altered gene expression, and oral tumor progression
Abstract
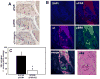
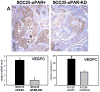
Oral squamous cell carcinoma (OSCC) has 50% 5-year survival rate, highlighting our limited understanding of the molecular events that contribute to disease progression. Microarray analyses of primary oral tumors have identified urinary-type plasminogen activator (uPA) and its receptor (uPAR) as key genes associated with human OSCC progression. The uPAR functions as both a proteinase receptor and an integrin ligand, modifying proteolysis, migration, integrin signaling, and cellular transcription. In the current study, uPAR expression levels were modified in OSCC cells followed by analysis of tumor growth in an in vivo orthotopic xenograft model and by transcriptional profiling. Overexpression of uPAR resulted in more infiltrative and less differentiated tumors, with ill-defined borders, cytologic atypia, and enhanced vascularity. Analysis of serial sections of both murine experimental tumors and microarrayed human OSCC showed a statistically significant association between uPAR and alpha(3) integrin colocalization in areas exhibiting extracellular signal-regulated kinase phosphorylation, suggesting that uPAR/alpha(3) integrin interaction potentiates extracellular signal-regulated kinase signaling in vivo. This is supported by cDNA microarray analysis, which showed differential expression of 148 genes (113 upregulated and 35 downregulated). Validation of gene expression changes in human OSCC using immunohistochemistry and quantitative real-time PCR showed increased growth factors, proteinases/inhibitors, and matrix components in uPAR-overexpressing tumors. Together, these results support a model wherein increased uPAR expression promotes alpha(3)beta(1) integrin association, resulting in increased mitogen-activated protein kinase signaling and transcriptional activation, leading to the formation of more aggressive tongue tumors. This combined approach has efficacy to identify additional biomarkers and/or prognostic indicators associated with aggressive human OSCC.
Figures





References
-
- Parkin DM, Pisani P, Ferlay J. Global cancer statistics. CA: A Cancer Journal for Clinicians. 1999:33–64. - PubMed
-
- Jemal A, Siegel R, Ward E, Murray T, Xu J, Smigal C, et al. Cancer statistics. CA: A Cancer Journal for Clinicians. 2006;56:106–130. - PubMed
-
- Gorsky M, Epstein BJ, Oakley C, Le ND, Hay J, Stevenson-Moore P. Carcinoma of the tongue: a case series analysis of clinical presentation, risk factors, staging and outcome. Oral Surg Oral Med Oral Pathol Oral Radio Endod. 2004;98:546–52. - PubMed
-
- Shiboski CH, Schmidt BL, Jordan RCK. Tongue and tonsil carcinoma: increasing trends in the US population ages 20–44 years. Cancer. 2005;103:1843–9. - PubMed
-
- Sano D, Myers JN. Metastasis of squamous cell carcinoma of the oral tongue. Cancer Metastasis Rev. 2007;26:645–62. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

