UpaH is a newly identified autotransporter protein that contributes to biofilm formation and bladder colonization by uropathogenic Escherichia coli CFT073
- PMID: 20145097
- PMCID: PMC2849410
- DOI: 10.1128/IAI.01010-09
UpaH is a newly identified autotransporter protein that contributes to biofilm formation and bladder colonization by uropathogenic Escherichia coli CFT073
Abstract
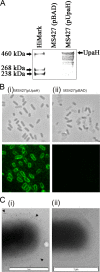
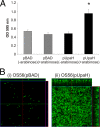
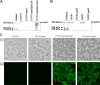
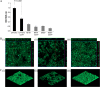
Escherichia coli is the primary cause of urinary tract infection (UTI) in the developed world. The major factors associated with virulence of uropathogenic E. coli (UPEC) are fimbrial adhesins, which mediate specific attachment to host receptors and trigger innate host responses. Another group of adhesins is represented by the autotransporter (AT) subgroup of proteins. In this study, we identified a new AT-encoding gene, termed upaH, present in a 6.5-kb unannotated intergenic region in the genome of the prototypic UPEC strain CFT073. Cloning and sequencing of the upaH gene from CFT073 revealed an intact 8.535-kb coding region, contrary to the published genome sequence. The upaH gene was widely distributed among a large collection of UPEC isolates as well as the E. coli Reference (ECOR) strain collection. Bioinformatic analyses suggest beta-helix as the predominant structure in the large N-terminal passenger (alpha) domain and a 12-strand beta-barrel for the C-terminal beta-domain of UpaH. We demonstrated that UpaH is expressed at the cell surface of CFT073 and promotes biofilm formation. In the mouse UTI model, deletion of the upaH gene in CFT073 and in two other UPEC strains did not significantly affect colonization of the bladder in single-challenge experiments. However, in competitive colonization experiments, CFT073 significantly outcompeted its upaH isogenic mutant strain in urine and the bladder.
Figures


References
-
- Anderson, G. G., J. J. Palermo, J. D. Schilling, R. Roth, J. Heuser, and S. J. Hultgren. 2003. Intracellular bacterial biofilm-like pods in urinary tract infections. Science 301:105-107. - PubMed
-
- Benz, I., and M. A. Schmidt. 1992. AIDA-I, the adhesin involved in diffuse adherence of the diarrhoeagenic Escherichia coli strain 2787 (O126:H27), is synthesized via a precursor molecule. Mol. Microbiol. 6:1539-1546. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources

